Identification of the hub genes associated with prostate cancer tumorigenesis
- PMID: 37251946
- PMCID: PMC10213256
- DOI: 10.3389/fonc.2023.1168772
Identification of the hub genes associated with prostate cancer tumorigenesis
Abstract
Introduction: Prostate cancer (PCa) is one of the most common malignant tumors of the male urogenital system; however, the underlying mechanisms remain largely unclear. This study integrated two cohort profile datasets to elucidate the potential hub genes and mechanisms in PCa.
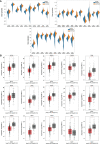
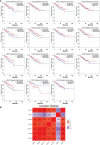
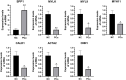
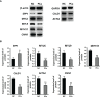
Methods and results: Gene expression profiles GSE55945 and GSE6919 were filtered from the Gene Expression Omnibus (GEO) database to obtain 134 differentially expressed genes (DEGs) (14 upregulated and 120 downregulated) in PCa. Gene Ontology and pathway enrichment were performed using the Database for Annotation, Visualization, and Integrated Discovery, showing that these DEGs were mainly involved in biological functions such as cell adhesion, extracellular matrix, migration, focal adhesion, and vascular smooth muscle contraction. The STRING database and Cytoscape tools were used to analyze protein-protein interactions and identify 15 hub candidate genes. Violin plot, boxplot, and prognostic curve analyses were performed using Gene Expression Profiling Interactive Analysis, which identified seven hub genes, including upregulated expressed SPP1 and downregulated expressed MYLK, MYL9, MYH11, CALD1, ACTA2, and CNN1 in PCa compared with normal tissue. Correlation analysis was performed using the OmicStudio tools, which showed that these hub genes were moderately to strongly correlated with each other. Finally, quantitative reverse transcription PCR and western blotting were performed to validate the hub genes, showing that the abnormal expression of the seven hub genes in PCa was consistent with the analysis results of the GEO database.
Discussion: Taken together, MYLK, MYL9, MYH11, CALD1, ACTA2, SPP1, and CNN1 are hub genes significantly associated with PCa occurrence. These genes are abnormally expressed, leading to the formation, proliferation, invasion, and migration of PCa cells and promoting tumor neovascularization. These genes may serve as potential biomarkers and therapeutic targets in patients with PCa.
Keywords: bioinformatics analysis; biomarkers; differentially expressed genes (DEGs); hub genes; prostate cancer.
Copyright © 2023 Zhu, Lin, Gao and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

