Nitrate enhances the secondary growth of storage roots in Panax ginseng
- PMID: 37252286
- PMCID: PMC10214138
- DOI: 10.1016/j.jgr.2022.05.009
Nitrate enhances the secondary growth of storage roots in Panax ginseng
Abstract
Background: Nitrogen (N) is an essential macronutrient for plant growth and development. To support agricultural production and enhance crop yield, two major N sources, nitrate and ammonium, are applied as fertilizers to the soil. Although many studies have been conducted on N uptake and signal transduction, the molecular genetic mechanisms of N-mediated physiological roles, such as the secondary growth of storage roots, remain largely unknown.
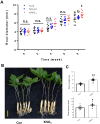
Methods: One-year-old P. ginseng seedlings treated with KNO3 were analyzed for the secondary growth of storage roots. The histological paraffin sections were subjected to bright and polarized light microscopic analysis. Genome-wide RNA-seq and network analysis were carried out to dissect the molecular mechanism of nitrate-mediated promotion of ginseng storage root thickening.
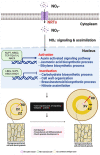
Results: Here, we report the positive effects of nitrate on storage root secondary growth in Panax ginseng. Exogenous nitrate supply to ginseng seedlings significantly increased the root secondary growth. Histological analysis indicated that the enhancement of root secondary growth could be attributed to the increase in cambium stem cell activity and the subsequent differentiation of cambium-derived storage parenchymal cells. RNA-seq and gene set enrichment analysis (GSEA) revealed that the formation of a transcriptional network comprising auxin, brassinosteroid (BR)-, ethylene-, and jasmonic acid (JA)-related genes mainly contributed to the secondary growth of ginseng storage roots. In addition, increased proliferation of cambium stem cells by a N-rich source inhibited the accumulation of starch granules in storage parenchymal cells.
Conclusion: Thus, through the integration of bioinformatic and histological tissue analyses, we demonstrate that nitrate assimilation and signaling pathways are integrated into key biological processes that promote the secondary growth of P. ginseng storage roots.
Keywords: Hormone; Nitrate; Panax ginseng; Root secondary growth.
© 2022 The Korean Society of Ginseng. Publishing services by Elsevier B.V.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Hu S.Y. The genusPanax (ginseng) in Chinese medicine. Econ Bot. 1976;30:11–28.
-
- Mahady G.B., Gyllenhall C., Fong H.H., Farnsworth N.R. Ginsengs: a review of safety and efficacy. Nutr Clin Care. 2000;3:90e101.
-
- Choi H.I., Waminal N.E., Park H.M., Kim N.H., Choi B.S., Park M., et al. Major repeat components covering one-third of the ginseng (Panax ginseng C.A. Meyer) genome and evidence for allotetraploidy. Plant J. 2014;77(6):906–916. - PubMed
LinkOut - more resources
Full Text Sources

