Learning perturbation-inducible cell states from observability analysis of transcriptome dynamics
- PMID: 37253722
- PMCID: PMC10229592
- DOI: 10.1038/s41467-023-37897-9
Learning perturbation-inducible cell states from observability analysis of transcriptome dynamics
Erratum in
-
Author Correction: Learning perturbation-inducible cell states from observability analysis of transcriptome dynamics.Nat Commun. 2024 Mar 6;15(1):2034. doi: 10.1038/s41467-024-46433-2. Nat Commun. 2024. PMID: 38448488 Free PMC article. No abstract available.
Abstract
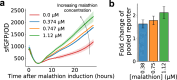
A major challenge in biotechnology and biomanufacturing is the identification of a set of biomarkers for perturbations and metabolites of interest. Here, we develop a data-driven, transcriptome-wide approach to rank perturbation-inducible genes from time-series RNA sequencing data for the discovery of analyte-responsive promoters. This provides a set of biomarkers that act as a proxy for the transcriptional state referred to as cell state. We construct low-dimensional models of gene expression dynamics and rank genes by their ability to capture the perturbation-specific cell state using a novel observability analysis. Using this ranking, we extract 15 analyte-responsive promoters for the organophosphate malathion in the underutilized host organism Pseudomonas fluorescens SBW25. We develop synthetic genetic reporters from each analyte-responsive promoter and characterize their response to malathion. Furthermore, we enhance malathion reporting through the aggregation of the response of individual reporters with a synthetic consortium approach, and we exemplify the library's ability to be useful outside the lab by detecting malathion in the environment. The engineered host cell, a living malathion sensor, can be optimized for use in environmental diagnostics while the developed machine learning tool can be applied to discover perturbation-inducible gene expression systems in the compendium of host organisms.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Bousse L. Whole cell biosensors. Sens. Actuat. B: Chem. 1996;34:270–275. doi: 10.1016/S0925-4005(96)01906-5. - DOI
-
- Song, Y. et al. Application of bacterial whole-cell biosensors in health. Handb. Cell Biosens. 945–961 (2022).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

