Antigen recognition detains CD8+ T cells at the blood-brain barrier and contributes to its breakdown
- PMID: 37253744
- PMCID: PMC10229608
- DOI: 10.1038/s41467-023-38703-2
Antigen recognition detains CD8+ T cells at the blood-brain barrier and contributes to its breakdown
Abstract
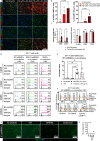
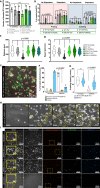
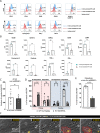
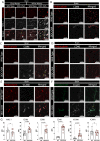
Blood-brain barrier (BBB) breakdown and immune cell infiltration into the central nervous system (CNS) are early hallmarks of multiple sclerosis (MS). High numbers of CD8+ T cells are found in MS lesions, and antigen (Ag) presentation at the BBB has been proposed to promote CD8+ T cell entry into the CNS. Here, we show that brain endothelial cells process and cross-present Ag, leading to effector CD8+ T cell differentiation. Under physiological flow in vitro, endothelial Ag presentation prevented CD8+ T cell crawling and diapedesis resulting in brain endothelial cell apoptosis and BBB breakdown. Brain endothelial Ag presentation in vivo was limited due to Ag uptake by CNS-resident macrophages but still reduced motility of Ag-specific CD8+ T cells within CNS microvessels. MHC class I-restricted Ag presentation at the BBB during neuroinflammation thus prohibits CD8+ T cell entry into the CNS and triggers CD8+ T cell-mediated focal BBB breakdown.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

