DNA topoisomerase 1 represses HIV-1 promoter activity through its interaction with a guanine quadruplex present in the LTR sequence
- PMID: 37254203
- PMCID: PMC10228017
- DOI: 10.1186/s12977-023-00625-8
DNA topoisomerase 1 represses HIV-1 promoter activity through its interaction with a guanine quadruplex present in the LTR sequence
Erratum in
-
Correction to: DNA topoisomerase 1 represses HIV-1 promoter activity through its interaction with a guanine quadruplex present in the LTR sequence.Retrovirology. 2023 Jul 11;20(1):12. doi: 10.1186/s12977-023-00627-6. Retrovirology. 2023. PMID: 37434132 Free PMC article. No abstract available.
Abstract
Background: Once integrated in the genome of infected cells, HIV-1 provirus is transcribed by the cellular transcription machinery. This process is regulated by both viral and cellular factors, which are necessary for an efficient viral replication as well as for the setting up of viral latency, leading to a repressed transcription of the integrated provirus.
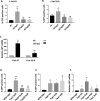
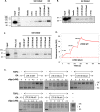
Results: In this study, we examined the role of two parameters in HIV-1 LTR promoter activity. We identified DNA topoisomerase1 (TOP1) to be a potent repressor of this promoter and linked this repression to its catalytic domain. Additionally, we confirmed the folding of a Guanine quadruplex (G4) structure in the HIV-1 promoter and its repressive effect. We demonstrated a direct interaction between TOP1 and this G4 structure, providing evidence of a functional relationship between the two repressive elements. Mutations abolishing G4 folding affected TOP1/G4 interaction and hindered G4-dependent inhibition of TOP1 catalytic activity in vitro. As a result, HIV-1 promoter activity was reactivated in a native chromatin environment. Lastly, we noticed an enrichment of predicted G4 sequences in the promoter of TOP1-repressed cellular genes.
Conclusions: Our results demonstrate the formation of a TOP1/G4 complex on the HIV-1 LTR promoter and its repressive effect on the promoter activity. They reveal the existence of a new mechanism of TOP1/G4-dependent transcriptional repression conserved between viral and human genes. This mechanism contrasts with the known property of TOP1 as global transcriptional activator and offers new perspectives for anti-cancer and anti-viral strategies.
Keywords: DNA topoisomerases; Guanine quadruplex; HIV-1 LTR promoter; HIV-1 latency; HIV-1 transcription; Host-virus interaction; Transcriptional regulation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare they have no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

