SARS-CoV-2 spike S2-specific neutralizing antibodies
- PMID: 37254830
- PMCID: PMC10274517
- DOI: 10.1080/22221751.2023.2220582
SARS-CoV-2 spike S2-specific neutralizing antibodies
Abstract
Since the onset of the coronavirus disease 2019 (COVID-19), numerous neutralizing antibodies (NAbs) against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been developed and authorized for emergency use to control the pandemic. Most COVID-19 therapeutic NAbs prevent the S1 subunit of the SARS-CoV-2 spike (S) protein from binding to the human host receptor. However, the emergence of SARS-CoV-2 immune escape variants, which possess frequent mutations on the S1 subunit, may render current NAbs ineffective. In contrast, the relatively conserved S2 subunit of the S protein can elicit NAbs with broader neutralizing potency against various SARS-CoV-2 variants. In this review, the binding specificity and functional features of SARS-CoV-2 NAbs targeting different domains of the S2 subunit are collectively discussed. The knowledge learned from the investigation of the S2-specific NAbs provides insights and potential strategies for developing antibody cocktail therapy and next-generation coronavirus vaccine.
Keywords: S2 subunit; SARS-CoV-2; antibody cocktail therapy; neutralizing antibody; spike protein.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

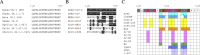

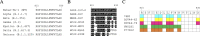

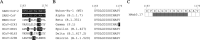
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
