To split or not to split: CASP15 targets and their processing into tertiary structure evaluation units
- PMID: 37254889
- PMCID: PMC10687315
- DOI: 10.1002/prot.26533
To split or not to split: CASP15 targets and their processing into tertiary structure evaluation units
Abstract
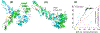
Processing of CASP15 targets into evaluation units (EUs) and assigning them to evolutionary-based prediction classes is presented in this study. The targets were first split into structural domains based on compactness and similarity to other proteins. Models were then evaluated against these domains and their combinations. The domains were joined into larger EUs if predictors' performance on the combined units was similar to that on individual domains. Alternatively, if most predictors performed better on the individual domains, then they were retained as EUs. As a result, 112 evaluation units were created from 77 tertiary structure prediction targets. The EUs were assigned to four prediction classes roughly corresponding to target difficulty categories in previous CASPs: TBM (template-based modeling, easy or hard), FM (free modeling), and the TBM/FM overlap category. More than a third of CASP15 EUs were attributed to the historically most challenging FM class, where homology or structural analogy to proteins of known fold cannot be detected.
Keywords: CASP15; evaluation units; protein domains; protein structure; protein structure prediction.
© 2023 Wiley Periodicals LLC.
Figures

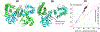

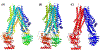
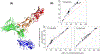

References
-
- Moult J, Pedersen JT, Judson R, Fidelis K. A large-scale experiment to assess protein structure prediction methods. Proteins 1995;23(3):ii–v. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

