CancerHERVdb: Human Endogenous Retrovirus (HERV) Expression Database for Human Cancer Accelerates Studies of the Retrovirome and Predictions for HERV-Based Therapies
- PMID: 37255431
- PMCID: PMC10308937
- DOI: 10.1128/jvi.00059-23
CancerHERVdb: Human Endogenous Retrovirus (HERV) Expression Database for Human Cancer Accelerates Studies of the Retrovirome and Predictions for HERV-Based Therapies
Abstract
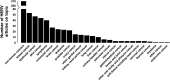
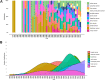
In this study, we sought to create a database summarizing the expression of human endogenous retroviruses (HERVs) in various human cancers. HERVs are suitable therapeutic targets due to their abundance in the human genome, overexpression in various malignancies, and involvement in various cancer pathways. We identified articles on HERVs from PubMed and then prescreened and automatically categorized them using the portable document format (PDF) data extractor (PDE) R package. We discovered 196 primary research articles with HERV expression data from cancer tissues or cancer cell lines. HERV RNA and protein expression was reported in brain, breast, cervical, colorectal, endocrine, gastrointestinal, kidney/renal/pelvis, liver, lung, genital, oral cavity, pharynx, ovary, pancreas, prostate, skin, testicular, urinary/bladder, and uterus cancers, leukemias, lymphomas, and myelomas. Additionally, we discovered reports of HERV RNA-only overexpression in soft tissue cancers including heart, thyroid, bone, and joint cancers. The CancerHERVdb database is hosted in the form of interactive visualizations of the expression data and a summary data table at https://erikstricker.shinyapps.io/cancerHERVdb/. The user can filter the findings according to cancer type, HERV family, HERV gene, or a combination thereof and easily export the results with the corresponding reference list. In our report, we provide examples of potential uses of the CancerHERVdb, such as identification of cancers suitable for off-target treatment with the multiple sclerosis-associated retrovirus (MSRV)-Env-targeting antibody GNbAC1 (now named temelimab) currently in phase 2b clinical trials for multiple sclerosis or the discovery of cancers overexpressing HERV-H long terminal repeat-associating 2 (HHLA2), a newly emerging immune checkpoint. In summary, the CancerHERVdb allows cross-study comparisons, encourages data exploration, and informs about potential off-target effects of HERV-targeting treatments. IMPORTANCE Human endogenous retroviruses (HERVs), which in the past have inserted themselves in various regions of the human genome, are to various degrees activated in virtually every cancer type. While a centralized naming system and resources summarizing HERV levels in cancers are lacking, the CancerHERVdb database provides a consolidated resource for cross-study comparisons, data exploration, and targeted searches of HERV activation. The user can access data extracted from hundreds of articles spanning 25 human cancer categories. Therefore, the CancerHERVdb database can aid in the identification of prognostic and risk markers, drivers of cancer, tumor-specific targets, multicancer spanning signals, and targets for immune therapies. Consequently, the CancerHERVdb database is of direct relevance for clinical as well as basic research.
Keywords: CancerHERVdb; HERV; cancer; database; expression; human endogenous retrovirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Serafino A, Balestrieri E, Pierimarchi P, Matteucci C, Moroni G, Oricchio E, Rasi G, Mastino A, Spadafora C, Garaci E, Vallebona PS. 2009. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp Cell Res 315:849–862. doi:10.1016/j.yexcr.2008.12.023. - DOI - PubMed
-
- Balestrieri E, Argaw-Denboba A, Gambacurta A, Cipriani C, Bei R, Serafino A, Sinibaldi-Vallebona P, Matteucci C. 2018. Human endogenous retrovirus K in the crosstalk between cancer cells microenvironment and plasticity: a new perspective for combination therapy. Front Microbiol 9:1448. doi:10.3389/fmicb.2018.01448. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

