Mapping the Kitchen Microbiota in Five European Countries Reveals a Set of Core Bacteria across Countries, Kitchen Surfaces, and Cleaning Utensils
- PMID: 37255441
- PMCID: PMC10304973
- DOI: 10.1128/aem.00267-23
Mapping the Kitchen Microbiota in Five European Countries Reveals a Set of Core Bacteria across Countries, Kitchen Surfaces, and Cleaning Utensils
Abstract
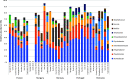
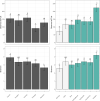
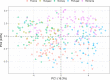
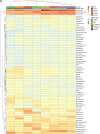
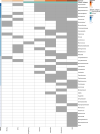
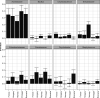
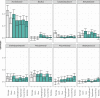
The residential kitchen is often heavily colonized by microbes originating from different sources, including food and human contact. Although a few studies have reported the bacterial composition in cleaning utensils and surface samples there is limited knowledge of the bacterial diversity across different sample types, households, and countries. As part of a large European study, we have identified the microbiota of 302 samples from cleaning utensils (sponges and cloths), kitchen surfaces (sinks, cutting boards, countertops, tap handles, and a pooled sample of other handles) in 74 households across 5 countries (France, Hungary, Norway, Portugal, and Romania). In total, 31 bacterial phyla were identified, with Proteobacteria, Firmicutes, Bacteroidota, and Actinobacteria being the most abundant. Despite large variations in households with respect to kitchen standards, kitchen practices, cleaning regimes, and diet and considerable differences in bacterial diversity between samples, eight bacterial genera/families commonly associated with environmental sources were identified in most samples and defined as a core microbiota: Acinetobacter, Pseudomonas, Enhydrobacter, Enterobacteriaceae, Psychrobacter, Chryseobacterium, Bacillus, and Staphylococcus. These genera/families were also among the bacteria with the highest relative abundance across all samples, in addition to Yersiniaceae, Kocuria, Pantoea, and Streptococcus. Taxa associated with potential pathogens and fecal indicators were low in abundance but broadly distributed throughout the households. The microbial composition of surface samples indicated that the microbial composition on kitchen surfaces is more characteristic for the particular country than the object type, while the microbiota of cleaning utensils was similar across countries but differed between types (sponge or cloth). IMPORTANCE There is limited knowledge of the characteristics, differences, and similarities of the bacterial composition in residential kitchens. Here, we report the microbiota of cleaning utensils (sponges and cloths) and five different surface samples in 74 households across five European countries. In addition to increasing the knowledge of the kitchen microbiota from many geographical areas, this study identified a core microbiota in European residential kitchens despite large variations in kitchen practices and kitchen design and standards across countries and households.
Keywords: 16S rRNA gene; Illumina; core microbiota; microbiome; sponges.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

