Hospital and community wastewater as a source of multidrug-resistant ESBL-producing Escherichia coli
- PMID: 37256105
- PMCID: PMC10225658
- DOI: 10.3389/fcimb.2023.1184081
Hospital and community wastewater as a source of multidrug-resistant ESBL-producing Escherichia coli
Abstract
Introduction: Hospitals and wastewater are recognized hot spots for the selection and dissemination of antibiotic-resistant bacteria to the environment, but the total participation of hospitals in the spread of nosocomial pathogens to municipal wastewater treatment plants (WWTPs) and adjacent rivers had not previously been revealed.
Methods: We used a combination of culturing and whole-genome sequencing to explore the transmission routes of Escherichia coli from hospitalized patients suffering from urinary tract infections (UTI) via wastewater to the environment. Samples were collected in two periods in three locations (A, B, and C) and cultured on selective antibiotic-enhanced plates.
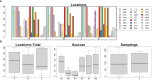
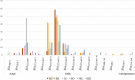
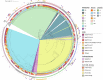
Results: In total, 408 E. coli isolates were obtained from patients with UTI (n=81), raw hospital sewage (n=73), WWTPs inflow (n=96)/outflow (n=106), and river upstream (n=21)/downstream (n=31) of WWTPs. The majority of the isolates produced extended-spectrum beta-lactamase (ESBL), mainly CTX-M-15, and showed multidrug resistance (MDR) profiles. Seven carbapenemase-producing isolates with GES-5 or OXA-244 were obtained in two locations from wastewater and river samples. Isolates were assigned to 74 different sequence types (ST), with the predominance of ST131 (n=80) found in all sources including rivers. Extraintestinal pathogenic lineages frequently found in hospital sewage (ST10, ST38, and ST69) were also found in river water. Despite generally high genetic diversity, phylogenetic analysis of ST10, ST295, and ST744 showed highly related isolates (SNP 0-18) from different sources, providing the evidence for the transmission of resistant strains through WWTPs to surface waters.
Discussion: Results of this study suggest that 1) UTI share a minor participation in hospitals wastewaters; 2) a high diversity of STs and phylogenetic groups in municipal wastewaters derive from the urban influence rather than hospitals; and 3) pathogenic lineages and bacteria with emerging resistance genotypes associated with hospitals spread into surface waters. Our study highlights the contribution of hospital and municipal wastewater to the transmission of ESBL- and carbapenemase-producing E. coli with MDR profiles to the environment.
Keywords: Escherichia coli; antibiotic resistance; beta-lactamases; wastewater; whole-genome sequencing.
Copyright © 2023 Davidova-Gerzova, Lausova, Sukkar, Nesporova, Nechutna, Vlkova, Chudejova, Krutova, Palkovicova, Kaspar and Dolejska.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

