Cx3cr1 controls kidney resident macrophage heterogeneity
- PMID: 37256130
- PMCID: PMC10225589
- DOI: 10.3389/fimmu.2023.1082078
Cx3cr1 controls kidney resident macrophage heterogeneity
Abstract
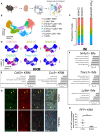
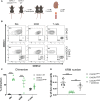
Kidney macrophages are comprised of both monocyte-derived and tissue resident populations; however, the heterogeneity of kidney macrophages and factors that regulate their heterogeneity are poorly understood. Herein, we performed single cell RNA sequencing (scRNAseq), fate mapping, and parabiosis to define the cellular heterogeneity of kidney macrophages in healthy mice. Our data indicate that healthy mouse kidneys contain four major subsets of monocytes and two major subsets of kidney resident macrophages (KRM) including a population with enriched Ccr2 expression, suggesting monocyte origin. Surprisingly, fate mapping data using the newly developed Ms4a3Cre Rosa Stopf/f TdT model indicate that less than 50% of Ccr2+ KRM are derived from Ly6chi monocytes. Instead, we find that Ccr2 expression in KRM reflects their spatial distribution as this cell population is almost exclusively found in the kidney cortex. We also identified Cx3cr1 as a gene that governs cortex specific accumulation of Ccr2+ KRM and show that loss of Ccr2+ KRM reduces the severity of cystic kidney disease in a mouse model where cysts are mainly localized to the kidney cortex. Collectively, our data indicate that Cx3cr1 regulates KRM heterogeneity and niche-specific disease progression.
Keywords: CCR2; CX3CR1; cystic kidney disease; fate mapping; kidney macrophages; macrophage heterogeneity; parabiosis; single cell RNA sequencing (scRNAseq).
Copyright © 2023 Yashchenko, Bland, Song, Ahmed, Sharp, Darby, Cordova, Smith, Lever, Li, Aloria, Khan, Maryam, Liu, Crowley, Jones, Zenewicz, George, Mrug, Crossman, Hopp, Stavrakis, Humphrey, Ginhoux and Zimmerman.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- P20 GM134973/GM/NIGMS NIH HHS/United States
- T32 AI007051/AI/NIAID NIH HHS/United States
- P30 DK090868/DK/NIDDK NIH HHS/United States
- R01 DK129255/DK/NIDDK NIH HHS/United States
- P30 DK074038/DK/NIDDK NIH HHS/United States
- I01 BX004232/BX/BLRD VA/United States
- P30 AR048311/AR/NIAMS NIH HHS/United States
- P30 DK079337/DK/NIDDK NIH HHS/United States
- U54 DK126087/DK/NIDDK NIH HHS/United States
- K01 DK119375/DK/NIDDK NIH HHS/United States
- K01 DK114164/DK/NIDDK NIH HHS/United States
- R01 DK097423/DK/NIDDK NIH HHS/United States
- P30 AI027767/AI/NIAID NIH HHS/United States
- I01 BX002298/BX/BLRD VA/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

