In vivo CRISPR screens reveal Serpinb9 and Adam2 as regulators of immune therapy response in lung cancer
- PMID: 37258521
- PMCID: PMC10232477
- DOI: 10.1038/s41467-023-38841-7
In vivo CRISPR screens reveal Serpinb9 and Adam2 as regulators of immune therapy response in lung cancer
Erratum in
-
Author Correction: In vivo CRISPR screens reveal Serpinb9 and Adam2 as regulators of immune therapy response in lung cancer.Nat Commun. 2023 Sep 5;14(1):5404. doi: 10.1038/s41467-023-41255-0. Nat Commun. 2023. PMID: 37670013 Free PMC article. No abstract available.
Abstract
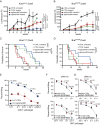
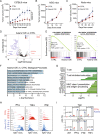
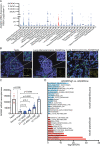
How the genetic landscape governs a tumor's response to immunotherapy remains poorly understood. To assess the immune-modulatory capabilities of 573 genes associated with altered cytotoxicity in human cancers, here we perform CRISPR/Cas9 screens directly in mouse lung cancer models. We recover the known immune evasion factors Stat1 and Serpinb9 and identify the cancer testis antigen Adam2 as an immune modulator, whose expression is induced by KrasG12D and further elevated by immunotherapy. Using loss- and gain-of-function experiments, we show that ADAM2 functions as an oncogene by restraining interferon and TNF cytokine signaling causing reduced presentation of tumor-associated antigens. ADAM2 also restricts expression of the immune checkpoint inhibitors PDL1, LAG3, TIGIT and TIM3 in the tumor microenvironment, which might explain why ex vivo expanded and adoptively transferred cytotoxic T-cells show enhanced cytotoxic efficacy in ADAM2 overexpressing tumors. Together, direct in vivo CRISPR/Cas9 screens can uncover genetic alterations that control responses to immunotherapies.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

