Engineered tRNAs suppress nonsense mutations in cells and in vivo
- PMID: 37258671
- PMCID: PMC10284701
- DOI: 10.1038/s41586-023-06133-1
Engineered tRNAs suppress nonsense mutations in cells and in vivo
Abstract
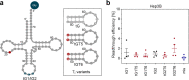
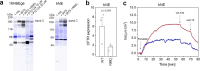
Nonsense mutations are the underlying cause of approximately 11% of all inherited genetic diseases1. Nonsense mutations convert a sense codon that is decoded by tRNA into a premature termination codon (PTC), resulting in an abrupt termination of translation. One strategy to suppress nonsense mutations is to use natural tRNAs with altered anticodons to base-pair to the newly emerged PTC and promote translation2-7. However, tRNA-based gene therapy has not yielded an optimal combination of clinical efficacy and safety and there is presently no treatment for individuals with nonsense mutations. Here we introduce a strategy based on altering native tRNAs into efficient suppressor tRNAs (sup-tRNAs) by individually fine-tuning their sequence to the physico-chemical properties of the amino acid that they carry. Intravenous and intratracheal lipid nanoparticle (LNP) administration of sup-tRNA in mice restored the production of functional proteins with nonsense mutations. LNP-sup-tRNA formulations caused no discernible readthrough at endogenous native stop codons, as determined by ribosome profiling. At clinically important PTCs in the cystic fibrosis transmembrane conductance regulator gene (CFTR), the sup-tRNAs re-established expression and function in cell systems and patient-derived nasal epithelia and restored airway volume homeostasis. These results provide a framework for the development of tRNA-based therapies with a high molecular safety profile and high efficacy in targeted PTC suppression.
© 2023. The Author(s).
Conflict of interest statement
Z.I., S.A., N.B. and M.D. are inventors on patents related to tRNA designs for PTC correction. Z.I. is also a scientific advisor for Tevard Biosciences. S.M.R. and G.J.T. are named on an unlicensed patent on the use of OCT for airway surface liquid measurements. S.M.R. is named on an unrelated patent on translational readthrough pharmacotherapy. E.J.S. is a non-voting board member of the Cystic Fibrosis Foundation. The LUNAR technology is proprietary to Arcturus Therapeutics. E.C.A., C.G.P.-G., R.M., B.M., K.K., M.A., K.J.-J.P., G.A., J.A.G., A.S., K.T., P.K., D.M. and P.C. are employees and have securities from Arcturus Therapeutics. The other authors declare no competing interests.
Figures












Comment in
-
Reading through stop codons with engineered tRNAs.Nat Rev Drug Discov. 2023 Aug;22(8):620. doi: 10.1038/d41573-023-00104-x. Nat Rev Drug Discov. 2023. PMID: 37369856 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

