A pan-influenza antibody inhibiting neuraminidase via receptor mimicry
- PMID: 37258672
- PMCID: PMC10266979
- DOI: 10.1038/s41586-023-06136-y
A pan-influenza antibody inhibiting neuraminidase via receptor mimicry
Erratum in
-
Author Correction: A pan-influenza antibody inhibiting neuraminidase via receptor mimicry.Nature. 2023 Jul;619(7970):E50. doi: 10.1038/s41586-023-06385-x. Nature. 2023. PMID: 37407829 Free PMC article. No abstract available.
Abstract
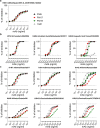
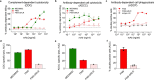
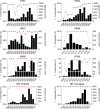
Rapidly evolving influenza A viruses (IAVs) and influenza B viruses (IBVs) are major causes of recurrent lower respiratory tract infections. Current influenza vaccines elicit antibodies predominantly to the highly variable head region of haemagglutinin and their effectiveness is limited by viral drift1 and suboptimal immune responses2. Here we describe a neuraminidase-targeting monoclonal antibody, FNI9, that potently inhibits the enzymatic activity of all group 1 and group 2 IAVs, as well as Victoria/2/87-like, Yamagata/16/88-like and ancestral IBVs. FNI9 broadly neutralizes seasonal IAVs and IBVs, including the immune-evading H3N2 strains bearing an N-glycan at position 245, and shows synergistic activity when combined with anti-haemagglutinin stem-directed antibodies. Structural analysis reveals that D107 in the FNI9 heavy chain complementarity-determinant region 3 mimics the interaction of the sialic acid carboxyl group with the three highly conserved arginine residues (R118, R292 and R371) of the neuraminidase catalytic site. FNI9 demonstrates potent prophylactic activity against lethal IAV and IBV infections in mice. The unprecedented breadth and potency of the FNI9 monoclonal antibody supports its development for the prevention of influenza illness by seasonal and pandemic viruses.
© 2023. The Author(s).
Conflict of interest statement
C.M., H.V.D., F.Z., K.H., C.W., J.d.I., A.M., N.C., A.D.M., K.B., D.D., S.V., A.C., B.G., A.E.P., S.S.Y., D.R.B., I.B., M.M., M.A.S., W.T.S., J.L.M., E.C., A.T., L.E.R., L.A.P., A.L., G.S., D.C. and M.S.P. are employees of and may hold shares in Vir Biotechnology. D.C., M.S.P., A.M., E.C., G.S., K.H., C.M. and E.F. are currently listed as inventors on multiple patent applications, which disclose the subject matter described in this paper. E.F., A.E.P., R.S. and H.W.V. are former employees of VIR Biotechnology. E.F. is currently an employee at and hold stocks in Amgen. Amgen provided no funding for this work. T.I.C. is a consultant for Vir Biotechnology and receives royalties from licensing of ISOLDE software used for molecular modelling. D.M.B. received research funding from VIR Biotechnology. L.A.P. is a former employee and shareholder of Regeneron Pharmaceuticals and is a member of the Scientific Advisory Board AI-driven structure-enabled antiviral platform (ASAP). Regeneron provided no funding for this work. H.W.V. is a founder of PierianDx and Casma Therapeutics. Neither company provided funding for this work. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential competing interest.
Figures












References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

