Structure and transport mechanism of the human calcium pump SPCA1
- PMID: 37258749
- PMCID: PMC10313705
- DOI: 10.1038/s41422-023-00827-x
Structure and transport mechanism of the human calcium pump SPCA1
Abstract
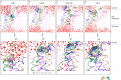
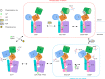
Secretory-pathway Ca2+-ATPases (SPCAs) play critical roles in maintaining Ca2+ homeostasis, but the exact mechanism of SPCAs-mediated Ca2+ transport remains unclear. Here, we determined six cryo-electron microscopy (cryo-EM) structures of human SPCA1 (hSPCA1) in a series of intermediate states, revealing a near-complete conformational cycle. With the aid of molecular dynamics simulations, these structures offer a clear structural basis for Ca2+ entry and release in hSPCA1. We found that hSPCA1 undergoes unique conformational changes during ATP binding and phosphorylation compared to other well-studied P-type II ATPases. In addition, we observed a conformational distortion of the Ca2+-binding site induced by the separation of transmembrane helices 4L and 6, unveiling a distinct Ca2+ release mechanism. Particularly, we determined a structure of the long-sought CaE2P state of P-type IIA ATPases, providing valuable insights into the Ca2+ transport cycle. Together, these findings enhance our understanding of Ca2+ transport by hSPCA1 and broaden our knowledge of P-type ATPases.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

