Normal cell cycle progression requires negative regulation of E2F1 by Groucho during S phase and its relief at G2 phase
- PMID: 37260146
- PMCID: PMC10281550
- DOI: 10.1242/dev.201041
Normal cell cycle progression requires negative regulation of E2F1 by Groucho during S phase and its relief at G2 phase
Abstract
The cell cycle depends on a sequence of steps that are triggered and terminated via the synthesis and degradation of phase-specific transcripts and proteins. Although much is known about how stage-specific transcription is activated, less is understood about how inappropriate gene expression is suppressed. Here, we demonstrate that Groucho, the Drosophila orthologue of TLE1 and other related human transcriptional corepressors, regulates normal cell cycle progression in vivo. We show that, although Groucho is expressed throughout the cell cycle, its activity is selectively inactivated by phosphorylation, except in S phase when it negatively regulates E2F1. Constitutive Groucho activity, as well as its depletion and the consequent derepression of e2f1, cause cell cycle phenotypes. Our results suggest that Cdk1 contributes to phase-specific phosphorylation of Groucho in vivo. We propose that Groucho and its orthologues play a role in the metazoan cell cycle that may explain the links between TLE corepressors and several types of human cancer.
Keywords: Drosophila; Cell cycle regulation; E2F1; Groucho; Protein phosphorylation; Repression.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures


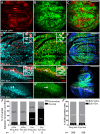
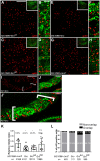
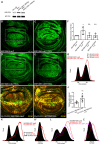
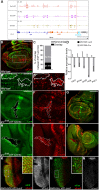

References
-
- Becker, K., Bluhm, A., Casas-Vila, N., Dinges, N., Dejung, M., Sayols, S., Kreutz, C., Roignant, J.-Y., Butter, F. and Legewie, S. (2018). Quantifying post-transcriptional regulation in the development of Drosophila melanogaster . Nat. Commun. 9, 4970. 10.1038/s41467-018-07455-9 - DOI - PMC - PubMed
-
- Bennett, J. D., Farlie, P. G. and Watson, R. J. (1996). E2F binding is required but not sufficient for repression of B-myb transcription in quiescent fibroblasts. Oncogene 13, 1073-1082. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

