Genomic Characterization of Theileria luwenshuni Strain Cheeloo
- PMID: 37260375
- PMCID: PMC10434005
- DOI: 10.1128/spectrum.00301-23
Genomic Characterization of Theileria luwenshuni Strain Cheeloo
Abstract
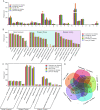
Theileria, a tick-borne intracellular protozoan, can cause infections of various livestock and wildlife around the world, posing a threat to veterinary health. Although more and more Theileria species have been identified, genomes have been available only from four Theileria species to date. Here, we assembled a whole genome of Theileria luwenshuni, an emerging Theileria, through next-generation sequencing of purified erythrocytes from the blood of a naturally infected goat. We designated it T. luwenshuni str. Cheeloo because its genome was assembled by the researchers at Cheeloo College of Medicine, Shandong University, China. The genome of T. lunwenshuni str. Cheeloo was the smallest in comparison with the other four Theileria species. T. luwenshuni str. Cheeloo possessed the fewest gene gains and gene family expansion. The protein count of each category was always comparable between T. luwenshuni str. Cheeloo and T. orientalis str. Shintoku in the Eukaryote Orthologs annotation, though there were remarkable differences in genome size. T. luwenshuni str. Cheeloo had lower counts than the other four Theileria species in most categories at level 3 of Gene Ontology annotation. Kyoto Encyclopedia of Genes and Genomes annotation revealed a loss of the c-Myb in T. luwenshuni str. Cheeloo. The infection rate of T. luwenshuni str. Cheeloo was up to 81.5% in a total of 54 goats from three flocks. The phylogenetic analyses based on both 18S rRNA and cox1 genes indicated that T. luwenshuni had relatively low diversity. The first characterization of the T. luwenshuni genome will promote better understanding of the emerging Theileria. IMPORTANCE Theileria has led to substantial economic losses in animal husbandry. Whole-genome sequencing data of the genus Theileria are currently limited, which has prohibited us from further understanding their molecular features. This work depicted whole-genome sequences of T. luwenshuni str. Cheeloo, an emerging Theileria species, and reported a high prevalence of T. luwenshuni str. Cheeloo infection in goats. The first assembly and characterization of T. luwenshuni genome will benefit exploring the infective and pathogenic mechanisms of the emerging Theileria to provide scientific basis for future control strategies of theileriosis.
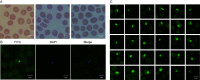
Keywords: FISH; Theileria luwenshuni strain Cheeloo; genomic characteristics; phylogenetic analysis; whole-genome sequence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
First molecular detection of Theileria luwenshuni from goats in Myanmar.Parasitol Res. 2018 Oct;117(10):3361-3364. doi: 10.1007/s00436-018-6073-6. Epub 2018 Sep 5. Parasitol Res. 2018. PMID: 30187170
-
Prevalence of Theileria infections in goats and sheep in southeastern China.Vet Parasitol. 2012 May 25;186(3-4):466-9. doi: 10.1016/j.vetpar.2011.11.066. Epub 2011 Nov 26. Vet Parasitol. 2012. PMID: 22178410
-
Migratory Gaddi sheep and goats as potential carriers of Theileria infection: a molecular survey.Trop Anim Health Prod. 2021 May 1;53(2):302. doi: 10.1007/s11250-021-02742-y. Trop Anim Health Prod. 2021. PMID: 33931794
-
First report of highly pathogenic Theileria luwenshuni in Sri Lanka: Are Jaffna sheep resistant to theileriosis?Vet Parasitol Reg Stud Reports. 2025 May;60:101250. doi: 10.1016/j.vprsr.2025.101250. Epub 2025 Mar 28. Vet Parasitol Reg Stud Reports. 2025. PMID: 40280673
-
Ovine theileriosis in China: a new look at an old story.Parasitol Res. 2007 Sep;101 Suppl 2:S191-5. doi: 10.1007/s00436-007-0689-2. Parasitol Res. 2007. PMID: 17823827 Review.
Cited by
-
Epidemiological and Molecular Characteristics of Piroplasmids and Anaplasma spp. in Tan Sheep, Ningxia, Northwest China.Transbound Emerg Dis. 2024 May 29;2024:2529855. doi: 10.1155/2024/2529855. eCollection 2024. Transbound Emerg Dis. 2024. PMID: 40303067 Free PMC article.
-
Comparative proteomics and structure-based approach to unravel the therapeutic drug target of Theileria species.J Genet Eng Biotechnol. 2025 Jun;23(2):100488. doi: 10.1016/j.jgeb.2025.100488. Epub 2025 Apr 10. J Genet Eng Biotechnol. 2025. PMID: 40390487 Free PMC article.
References
-
- Pal A, Chakravarty AK. 2020. Chapter 2 - major diseases of livestock and poultry and problems encountered in controlling them. Genetics and Breeding for Disease Resistance of Livestock:11–83.
-
- Tretina K, Pelle R, Orvis J, Gotia HT, Ifeonu OO, Kumari P, Palmateer NC, Iqbal SBA, Fry LM, Nene VM, Daubenberger CA, Bishop RP, Silva JC. 2020. Re-annotation of the Theileria parva genome refines 53% of the proteome and uncovers essential components of N-glycosylation, a conserved pathway in many organisms. BMC Genomics 21:279. doi:10.1186/s12864-020-6683-0. - DOI - PMC - PubMed
-
- Hayashida K, Hara Y, Abe T, Yamasaki C, Toyoda A, Kosuge T, Suzuki Y, Sato Y, Kawashima S, Katayama T, Wakaguri H, Inoue N, Homma K, Tada-Umezaki M, Yagi Y, Fujii Y, Habara T, Kanehisa M, Watanabe H, Ito K, Gojobori T, Sugawara H, Imanishi T, Weir W, Gardner M, Pain A, Shiels B, Hattori M, Nene V, Sugimoto C. 2012. Comparative genome analysis of three eukaryotic parasites with differing abilities to transform leukocytes reveals key mediators of Theileria-induced leukocyte transformation. mBio 3:e00204-12–e00212. doi:10.1128/mBio.00204-12. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

