Escherichia coli ST2797 Is Abundant in Wastewater and Might Be a Novel Emerging Extended-Spectrum Beta-Lactamase E. coli
- PMID: 37260395
- PMCID: PMC10434162
- DOI: 10.1128/spectrum.04486-22
Escherichia coli ST2797 Is Abundant in Wastewater and Might Be a Novel Emerging Extended-Spectrum Beta-Lactamase E. coli
Abstract
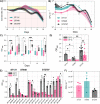
The increasing prevalence of antibiotic-resistant bacteria is an emerging threat to global health. The analysis of antibiotic-resistant enterobacteria in wastewater can indicate the prevalence and spread of certain clonal groups of multiresistant bacteria. In a previous study of Escherichia coli that were isolated from a pump station in Norway over 15 months, we found a recurring E. coli clone that was resistant to trimethoprim, ampicillin, and tetracycline in 201 of 3,123 analyzed isolates (6.1%). 11 representative isolates were subjected to whole-genome sequencing and were found to belong to the MLST ST2797 E. coli clone with plasmids carrying resistance genes, including blaTEM-1B, sul2, dfrA7, and tetB. A phenotypic comparison of the ST2797 isolates with the uropathogenic ST131 and ST648 that were repeatedly identified in the same wastewater samples revealed that the ST2797 isolates exhibited a comparable capacity for temporal survival in wastewater, greater biofilm formation, and similar potential for the colonization of mammalian epithelial cells. ST2797 has been isolated from humans and has been found to carry extended spectrum β-lactamase (ESBL) genes in other studies, suggesting that this clonal type is an emerging ESBL E. coli. Collectively, these findings show that ST2797 was more ubiquitous in the studied wastewater than were the infamous ST131 and ST648 and that ST2797 may have similar abilities to survive in the environment and cause infections in humans. IMPORTANCE The incidence of drug-resistant bacteria found in the environment is increasing together with the levels of antibiotic-resistant bacteria that cause infections. The COVID-19 pandemic has shed new light on the importance of monitoring emerging threats and finding early warning systems. Therefore, to mitigate the antimicrobial resistance burden, the monitoring and early identification of antibiotic-resistant bacteria in hot spots, such as wastewater treatment plants, are required to combat the occurrence and spread of antibiotic-resistant bacteria. Here, we applied a PhenePlate system as a phenotypic screening method for genomic surveillance and discovered a dominant and persistent E. coli clone ST2797 with a multidrug resistance pattern and equivalent phenotypic characteristics to those of the major pandemic lineages, namely, ST131 and ST648, which frequently carry ESBL genes. This study highlights the continuous surveillance and report of multidrug resistant bacteria with the potential to spread in One Health settings.
Keywords: Escherichia coli; PhenePlate; ST2797; bacterial survival; biochemical fingerprinting; biofilm; genomics; multidrug resistant; multidrug resistant bacteria; persistent clones; urban wastewater.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I, Fatta-Kassinos D. 2013. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447:345–360. doi:10.1016/j.scitotenv.2013.01.032. - DOI - PubMed
-
- Paulshus E, Thorell K, Guzman-Otazo J, Joffre E, Colque P, Kühn I, Möllby R, Sørum H, Sjöling Å. 2019. Repeated isolation of extended-spectrum-β-lactamase-positive Escherichia coli sequence types 648 and 131 from community wastewater indicates that sewage systems are important sources of emerging clones of antibiotic-resistant bacteria. Antimicrob Agents Chemother 63. doi:10.1128/AAC.00823-19. - DOI - PMC - PubMed
-
- Paulshus E, Kühn I, Möllby R, Colque P, O'Sullivan K, Midtvedt T, Lingaas E, Holmstad R, Sørum H. 2019. Diversity and antibiotic resistance among Escherichia coli populations in hospital and community wastewater compared to wastewater at the receiving urban treatment plant. Water Res 161:232–241. doi:10.1016/j.watres.2019.05.102. - DOI - PubMed
-
- Rudbeck E, Sharma A, Kirangwa J, Hu Y, Álvarez-Carretero S, Fredrik B, Thorell K. 2017. 2021. BACTpipe: bacterial whole genome sequence assembly and annotation pipeline. https://github.com/ctmrbio/BACTpipe. doi:10.5281/zenodo.4742358. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

