Dysregulated early transcriptional signatures linked to mast cell and interferon responses are implicated in COVID-19 severity
- PMID: 37261339
- PMCID: PMC10229044
- DOI: 10.3389/fimmu.2023.1166574
Dysregulated early transcriptional signatures linked to mast cell and interferon responses are implicated in COVID-19 severity
Abstract
Background: Dysregulated immune responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection are thought to underlie the progression of coronavirus disease 2019 (COVID-19) to severe disease. We sought to determine whether early host immune-related gene expression could predict clinical progression to severe disease.
Methods: We analysed the expression of 579 immunological genes in peripheral blood mononuclear cells taken early after symptom onset using the NanoString nCounter and compared SARS-CoV-2 negative controls with SARS-CoV-2 positive subjects with mild (SARS+ Mild) and Moderate/Severe disease to evaluate disease outcomes. Biobanked plasma samples were also assessed for type I (IFN-α2a and IFN-β), type II (IFN-γ) and type III (IFN-λ1) interferons (IFNs) as well as 10 additional cytokines using multiplex immunoassays.
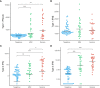
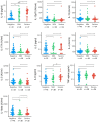
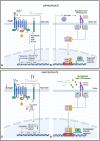
Results: We identified 19 significantly deregulated genes in 62 SARS-CoV-2 positive subject samples within 5 days of symptom onset and 58 SARS-CoV-2 negative controls and found that type I interferon (IFN) signalling (MX1, IRF7, IFITM1, IFI35, STAT2, IRF4, PML, BST2, STAT1) and genes encoding proinflammatory cytokines (TNF, TNFSF4, PTGS2 and IL1B) were upregulated in both SARS+ groups. Moreover, we found that FCER1, involved in mast cell activation, was upregulated in the SARS+ Mild group but significantly downregulated in the SARS+ Moderate/Severe group. In both SARS+ groups we discovered elevated interferon type I IFN-α2a, type II IFN and type III IFN λ1 plasma levels together with higher IL-10 and IL-6. These results indicate that those with moderate or severe disease are characterised by deficiencies in a mast cell response together with IFN hyper-responsiveness, suggesting that early host antiviral immune responses could be a cause and not a consequence of severe COVID-19.
Conclusions: This study suggests that early host immune responses linking defects in mast cell activation with host interferon responses correlates with more severe outcomes in COVID-19. Further characterisation of this pathway could help inform better treatment for vulnerable individuals.
Keywords: COVID-19; SARS-CoV-2; gene expression; interferon; mast cells; plasma biomarkers.
Copyright © 2023 MacCann, Leon, Gonzalez, Carr, Feeney, Yousif, Cotter, de Barra, Sadlier, Doran and Mallon.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

