Neutrophil extracellular traps and extracellular histones potentiate IL-17 inflammation in periodontitis
- PMID: 37261457
- PMCID: PMC10236943
- DOI: 10.1084/jem.20221751
Neutrophil extracellular traps and extracellular histones potentiate IL-17 inflammation in periodontitis
Abstract
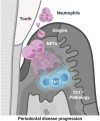
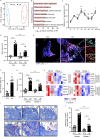
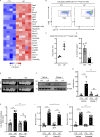
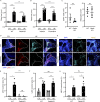
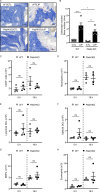
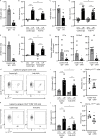
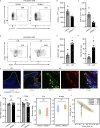
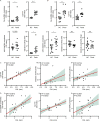
Neutrophil infiltration is a hallmark of periodontitis, a prevalent oral inflammatory condition in which Th17-driven mucosal inflammation leads to destruction of tooth-supporting bone. Herein, we document that neutrophil extracellular traps (NETs) are early triggers of pathogenic inflammation in periodontitis. In an established animal model, we demonstrate that neutrophils infiltrate the gingival oral mucosa at early time points after disease induction and expel NETs to trigger mucosal inflammation and bone destruction in vivo. Investigating mechanisms by which NETs drive inflammatory bone loss, we find that extracellular histones, a major component of NETs, trigger upregulation of IL-17/Th17 responses, and bone destruction. Importantly, human findings corroborate our experimental work. We document significantly increased levels of NET complexes and extracellular histones bearing classic NET-associated posttranslational modifications, in blood and local lesions of severe periodontitis patients, in the absence of confounding disease. Our findings suggest a feed-forward loop in which NETs trigger IL-17 immunity to promote immunopathology in a prevalent human inflammatory disease.
© 2023 Moutsopoulos et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures


Comment in
-
NETs implicated in periodontitis-associated bone loss.Nat Rev Rheumatol. 2023 Aug;19(8):463. doi: 10.1038/s41584-023-01000-9. Nat Rev Rheumatol. 2023. PMID: 37430108 No abstract available.
-
NET histones inflame periodontitis.J Exp Med. 2023 Sep 4;220(9):e20230783. doi: 10.1084/jem.20230783. Epub 2023 Aug 8. J Exp Med. 2023. PMID: 37552469 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

