Emergence, spread and characterisation of the SARS-CoV-2 variant B.1.640 circulating in France, October 2021 to February 2022
- PMID: 37261732
- PMCID: PMC10236926
- DOI: 10.2807/1560-7917.ES.2023.28.22.2200671
Emergence, spread and characterisation of the SARS-CoV-2 variant B.1.640 circulating in France, October 2021 to February 2022
Abstract
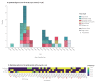
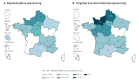
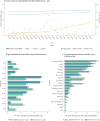
BackgroundSuccessive epidemic waves of COVID-19 illustrated the potential of SARS-CoV-2 variants to reshape the pandemic. Detecting and characterising emerging variants is essential to evaluate their public health impact and guide implementation of adapted control measures.AimTo describe the detection of emerging variant, B.1.640, in France through genomic surveillance and present investigations performed to inform public health decisions.MethodsIdentification and monitoring of SARS-CoV-2 variant B.1.640 was achieved through the French genomic surveillance system, producing 1,009 sequences. Additional investigation of 272 B.1.640-infected cases was performed between October 2021 and January 2022 using a standardised questionnaire and comparing with Omicron variant-infected cases.ResultsB.1.640 was identified in early October 2021 in a school cluster in Bretagne, later spreading throughout France. B.1.640 was detected at low levels at the end of SARS-CoV-2 Delta variant's dominance and progressively disappeared after the emergence of the Omicron (BA.1) variant. A high proportion of investigated B.1.640 cases were children aged under 14 (14%) and people over 60 (27%) years, because of large clusters in these age groups. B.1.640 cases reported previous SARS-CoV-2 infection (4%), anosmia (32%) and ageusia (34%), consistent with data on pre-Omicron SARS-CoV-2 variants. Eight percent of investigated B.1.640 cases were hospitalised, with an overrepresentation of individuals aged over 60 years and with risk factors.ConclusionEven though B.1.640 did not outcompete the Delta variant, its importation and continuous low-level spread raised concerns regarding its public health impact. The investigations informed public health decisions during the time that B.1.640 was circulating.
Keywords: B.1.640; COVID-19; SARS-CoV-2; emerging variant; genomic surveillance; whole genome sequencing.
Conflict of interest statement
Figures
References
-
- World Health Organization (WHO). Weekly epidemiological update on COVID-19 - 4 January 2023, edition 124. Geneva: WHO; 2022. Available from: https://www.who.int/publications/m/item/weekly-epidemiological-update-on...
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
