Macrophage-endothelial cell crosstalk orchestrates neutrophil recruitment in inflamed mucosa
- PMID: 37261911
- PMCID: PMC10378177
- DOI: 10.1172/JCI170733
Macrophage-endothelial cell crosstalk orchestrates neutrophil recruitment in inflamed mucosa
Abstract
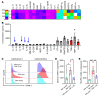
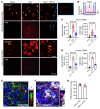
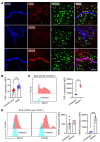
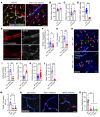
Neutrophil (PMN) mobilization to sites of insult is critical for host defense and requires transendothelial migration (TEM). TEM involves several well-studied sequential adhesive interactions with vascular endothelial cells (ECs); however, what initiates or terminates this process is not well-understood. Here, we describe what we believe to be a new mechanism where vessel-associated macrophages through localized interactions primed EC responses to form ICAM-1 "hot spots" to support PMN TEM. Using real-time intravital microscopy of LPS-inflamed intestines in CX3CR1-EGFP macrophage-reporter mice, complemented by whole-mount tissue imaging and flow cytometry, we found that macrophage vessel association is critical for the initiation of PMN-EC adhesive interactions, PMN TEM, and subsequent accumulation in the intestinal mucosa. Anti-colony stimulating factor 1 receptor Ab-mediated macrophage depletion in the lamina propria and at the vessel wall resulted in elimination of ICAM-1 hot spots impeding PMN-EC interactions and TEM. Mechanistically, the use of human clinical specimens, TNF-α-KO macrophage chimeras, TNF-α/TNF receptor (TNF-α/TNFR) neutralization, and multicellular macrophage-EC-PMN cocultures revealed that macrophage-derived TNF-α and EC TNFR2 axis mediated this regulatory mechanism and was required for PMN TEM. As such, our findings identified clinically relevant mechanisms by which macrophages regulate PMN trafficking in inflamed mucosa.
Keywords: Cell Biology; Cell migration/adhesion; Inflammation; Macrophages; Neutrophils.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

