Reversing pathological cell states: the road less travelled can extend the therapeutic horizon
- PMID: 37263821
- PMCID: PMC10593090
- DOI: 10.1016/j.tcb.2023.04.004
Reversing pathological cell states: the road less travelled can extend the therapeutic horizon
Abstract
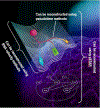
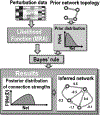
Acquisition of omics data advances at a formidable pace. Yet, our ability to utilize these data to control cell phenotypes and design interventions that reverse pathological states lags behind. Here, we posit that cell states are determined by core networks that control cell-wide networks. To steer cell fate decisions, core networks connecting genotype to phenotype must be reconstructed and understood. A recent method, cell state transition assessment and regulation (cSTAR), applies perturbation biology to quantify causal connections and mechanistically models how core networks influence cell phenotypes. cSTAR models are akin to digital cell twins enabling us to purposefully convert pathological states back to physiologically normal states. While this capability has a range of applications, here we discuss reverting oncogenic transformation.
Keywords: cell state transition assessment and regulation method; control of cell state transitions and fate decisions; digital cell twins; omics data.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests Patent application (No. UK2107576.7) related to this work was filed by the authors.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

