Rio1 downregulates centromeric RNA levels to promote the timely assembly of structurally fit kinetochores
- PMID: 37263996
- PMCID: PMC10235086
- DOI: 10.1038/s41467-023-38920-9
Rio1 downregulates centromeric RNA levels to promote the timely assembly of structurally fit kinetochores
Abstract
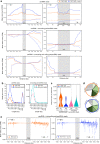
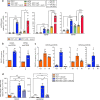
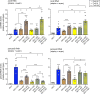
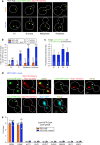
Kinetochores assemble on centromeres via histone H3 variant CENP-A and low levels of centromere transcripts (cenRNAs). The latter are ensured by the downregulation of RNA polymerase II (RNAPII) activity, and cenRNA turnover by the nuclear exosome. Using S. cerevisiae, we now add protein kinase Rio1 to this scheme. Yeast cenRNAs are produced either as short (median lengths of 231 nt) or long (4458 nt) transcripts, in a 1:1 ratio. Rio1 limits their production by reducing RNAPII accessibility and promotes cenRNA degradation by the 5'-3'exoribonuclease Rat1. Rio1 similarly curtails the concentrations of noncoding pericenRNAs. These exist as short transcripts (225 nt) at levels that are minimally two orders of magnitude higher than the cenRNAs. In yeast depleted of Rio1, cen- and pericenRNAs accumulate, CEN nucleosomes and kinetochores misform, causing chromosome instability. The latter phenotypes are also observed with human cells lacking orthologue RioK1, suggesting that CEN regulation by Rio1/RioK1 is evolutionary conserved.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

