HLA class I signal peptide polymorphism determines the level of CD94/NKG2-HLA-E-mediated regulation of effector cell responses
- PMID: 37264229
- PMCID: PMC10690437
- DOI: 10.1038/s41590-023-01523-z
HLA class I signal peptide polymorphism determines the level of CD94/NKG2-HLA-E-mediated regulation of effector cell responses
Abstract
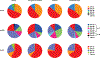
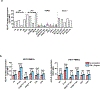
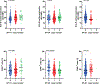
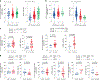
Human leukocyte antigen (HLA)-E binds epitopes derived from HLA-A, HLA-B, HLA-C and HLA-G signal peptides (SPs) and serves as a ligand for CD94/NKG2A and CD94/NKG2C receptors expressed on natural killer and T cell subsets. We show that among 16 common classical HLA class I SP variants, only 6 can be efficiently processed to generate epitopes that enable CD94/NKG2 engagement, which we term 'functional SPs'. The single functional HLA-B SP, known as HLA-B/-21M, induced high HLA-E expression, but conferred the lowest receptor recognition. Consequently, HLA-B/-21M SP competes with other SPs for providing epitope to HLA-E and reduces overall recognition of target cells by CD94/NKG2A, calling for reassessment of previous disease models involving HLA-B/-21M. Genetic population data indicate a positive correlation between frequencies of functional SPs in humans and corresponding cytomegalovirus mimics, suggesting a means for viral escape from host responses. The systematic, quantitative approach described herein will facilitate development of prediction algorithms for accurately measuring the impact of CD94/NKG2-HLA-E interactions in disease resistance/susceptibility.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures



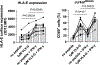





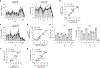

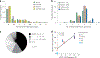

References
-
- McMahon CW & Raulet DH Expression and function of NK cell receptors in CD8+ T cells. Curr. Opin. Immunol. 13, 465–470 (2001). - PubMed
-
- Borrego F, Ulbrecht M, Weiss EH, Coligan JE & Brooks AG Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J. Exp. Med. 187, 813–818 (1998). - PMC - PubMed
-
- Braud VM et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799 (1998). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

