The glycine receptor alpha 3 subunit mRNA expression shows sex-dependent differences in the adult mouse brain
- PMID: 37264306
- PMCID: PMC10233971
- DOI: 10.1186/s12868-023-00800-9
The glycine receptor alpha 3 subunit mRNA expression shows sex-dependent differences in the adult mouse brain
Abstract
Background: The glycinergic system plays an important inhibitory role in the mouse central nervous system, where glycine controls the excitability of spinal itch- and pain-mediating neurons. Impairments of the glycine receptors can cause motor and sensory deficits. Glycine exerts inhibition through interaction with ligand-gated ion channels composed of alpha and beta subunits. We have investigated the mRNA expression of the glycine receptor alpha 3 (Glra3) subunit in the nervous system as well as in several peripheral organs of female and male mice.
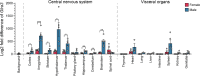
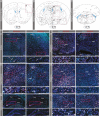
Results: Single-cell RNA sequencing (scRNA-seq) data analysis on the Zeisel et al. (2018) dataset indicated widespread but low expression of Glra3 in vesicular glutamate transporter 2 (Vglut2, Slc17a6) positive and vesicular inhibitory amino acid transporter (Viaat, Slc32a1)positive neurons of the mouse central nervous system. Highest occurrence of Glra3 expression was identified in the cortex, amygdala, and striatal regions, as well as in the hypothalamus, brainstem and spinal cord. Bulk quantitative real-time-PCR (qRT-PCR) analysis demonstrated Glra3 expression in cortex, amygdala, striatum, hypothalamus, thalamus, pituitary gland, hippocampus, cerebellum, brainstem, and spinal cord. Additionally, male mice expressed higher levels of Glra3 in all investigated brain areas compared with female mice. Lastly, RNAscope spatially validated Glra3 expression in the areas indicated by the single-cell and bulk analyses. Moreover, RNAscope analysis confirmed co-localization of Glra3 with Slc17a6 or Slc32a1 in the central nervous system areas suggested from the single-cell data.
Conclusions: Glra3 expression is low but widespread in the mouse central nervous system. Clear sex-dependent differences have been identified, indicating higher levels of Glra3 in several telencephalic and diencephalic areas, as well as in cerebellum and brainstem, in male mice compared with female mice.
Keywords: Brain; Glra3; Glycine; Mice; Sex-dependent differences; Spinal cord.
© 2023. The Author(s).
Conflict of interest statement
The authors state no conflict of interest.
Figures




Similar articles
-
Spinal Glycine Receptor Alpha 3 Cells Communicate Sensations of Chemical Itch in Hairy Skin.J Neurosci. 2024 May 8;44(19):e1585232024. doi: 10.1523/JNEUROSCI.1585-23.2024. J Neurosci. 2024. PMID: 38553047 Free PMC article.
-
The mRNA expression profile of glycine receptor subunits alpha 1, alpha 2, alpha 4 and beta in female and male mice.Mol Cell Neurosci. 2024 Dec;131:103976. doi: 10.1016/j.mcn.2024.103976. Epub 2024 Nov 22. Mol Cell Neurosci. 2024. PMID: 39580061
-
Presynaptic facilitation of glycinergic mIPSC is reduced in mice lacking α3 glycine receptor subunits.Neuroscience. 2016 Apr 21;320:1-7. doi: 10.1016/j.neuroscience.2016.01.063. Epub 2016 Feb 3. Neuroscience. 2016. PMID: 26851771
-
Developmental Formation of the GABAergic and Glycinergic Networks in the Mouse Spinal Cord.Int J Mol Sci. 2022 Jan 13;23(2):834. doi: 10.3390/ijms23020834. Int J Mol Sci. 2022. PMID: 35055019 Free PMC article. Review.
-
Glycinergic transmission.Cell Tissue Res. 2006 Nov;326(2):535-40. doi: 10.1007/s00441-006-0261-x. Epub 2006 Jun 29. Cell Tissue Res. 2006. PMID: 16807723 Review.
Cited by
-
Spinal Glycine Receptor Alpha 3 Cells Communicate Sensations of Chemical Itch in Hairy Skin.J Neurosci. 2024 May 8;44(19):e1585232024. doi: 10.1523/JNEUROSCI.1585-23.2024. J Neurosci. 2024. PMID: 38553047 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

