Pervasive tandem duplications and convergent evolution shape coral genomes
- PMID: 37264421
- PMCID: PMC10236652
- DOI: 10.1186/s13059-023-02960-7
Pervasive tandem duplications and convergent evolution shape coral genomes
Abstract
Background: Over the last decade, several coral genomes have been sequenced allowing a better understanding of these symbiotic organisms threatened by climate change. Scleractinian corals are reef builders and are central to coral reef ecosystems, providing habitat to a great diversity of species.
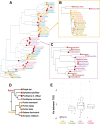
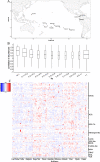
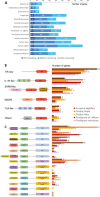
Results: In the frame of the Tara Pacific expedition, we assemble two coral genomes, Porites lobata and Pocillopora cf. effusa, with vastly improved contiguity that allows us to study the functional organization of these genomes. We annotate their gene catalog and report a relatively higher gene number than that found in other public coral genome sequences, 43,000 and 32,000 genes, respectively. This finding is explained by a high number of tandemly duplicated genes, accounting for almost a third of the predicted genes. We show that these duplicated genes originate from multiple and distinct duplication events throughout the coral lineage. They contribute to the amplification of gene families, mostly related to the immune system and disease resistance, which we suggest to be functionally linked to coral host resilience.
Conclusions: At large, we show the importance of duplicated genes to inform the biology of reef-building corals and provide novel avenues to understand and screen for differences in stress resilience.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests. JMA received travel and accommodation expenses to speak at Oxford Nanopore Technologies conferences.
Figures





References
-
- World Conservation Monitoring Centre, United Nations Environment Programme. World atlas of coral reefs. [Internet]. 2001. [quote: 2023, May] Available from: https://wedocs.unep.org/20.500.11822/30238
-
- Moberg F, Folke C. Ecological goods and services of coral reef ecosystems. Ecol Econ. 1999;29:215–233. doi: 10.1016/S0921-8009(99)00009-9. - DOI
-
- Pogoreutz C, et al. The coral holobiont highlights the dependence of cnidarian animal hosts on their associated microbes. in Cellular Dialogues in the Holobiont (CRC Press, 2020).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

