Combined analysis of transcriptomics and metabolomics revealed complex metabolic genes for diterpenoids biosynthesis in different organs of Anoectochilus roxburghii
- PMID: 37265764
- PMCID: PMC10230631
- DOI: 10.1016/j.chmed.2022.11.002
Combined analysis of transcriptomics and metabolomics revealed complex metabolic genes for diterpenoids biosynthesis in different organs of Anoectochilus roxburghii
Abstract
Objective: Diterpenoids with a wide variety of biological activities from Anoectochilus roxburghii, a precious medicinal plant, are important active components. However, due to the lack of genetic information on the metabolic process of diterpenoids in A. roxburghii, the genes involved in the molecular regulation mechanism of diterpenoid metabolism are still unclear. This study revealed the complex metabolic genes for diterpenoids biosynthesis in different organs of A. roxburghii by combining analysis of transcriptomics and metabolomics.
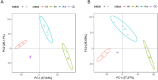
Methods: The differences in diterpenoid accumulation in roots, stems and leaves of A. roxburghii were analyzed by metabonomic analysis, and its metabolic gene information was obtained by transcriptome sequencing. Then, the molecular mechanism of differential diterpenoid accumulation in different organs of A. roxburghii was analyzed from the perspective of gene expression patterns.
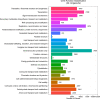
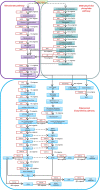
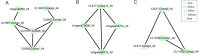
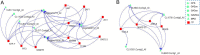
Results: A total of 296 terpenoid metabolites were identified in the five terpenoid metabolic pathways in A. roxburghii. There were 38, 34, and 18 diterpenoids with different contents between roots and leaves, between leaves and stems, and between roots and stems, respectively. Twenty-nine metabolic enzyme genes with 883 unigenes in the diterpenoid synthesis process were identified, and the DXS and FDPS in the terpenoid backbone biosynthesis stage and CPA, GA20ox, GA3ox, GA2ox, and MAS in the diterpenoid biosynthesis stage were predicted to be the key metabolic enzymes for the accumulation of diterpenoids. In addition, 14 key transcription factor coding genes were predicted to be involved in the regulation of the diterpenoid biosynthesis. The expression of genes such as GA2ox, MAS, CPA, GA20ox and GA3ox might be activated by some of the 14 transcription factors. The transcription factor NTF-Y and PRE6 were predicted to be the most important transcription factors.
Conclusion: This study determined 29 metabolic enzyme genes and predicted 14 transcription factors involved in the molecular regulation mechanism of diterpenoid metabolism in A. roxburghii, which provided a reference for the further study of the molecular regulation mechanism of the accumulation of diterpenoids in different organs of A. roxburghii.
Keywords: Anoectochilus roxburghii (Wall.) Lindl.; diterpenoid metabolic pathway; leaves; metabolomics; roots; stems; transcriptome.
© 2022 Tianjin Press of Chinese Herbal Medicines. Published by ELSEVIER B.V.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
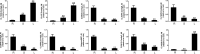
Similar articles
-
Comparative transcriptome analysis of roots, stems and leaves of Isodon amethystoides reveals candidate genes involved in Wangzaozins biosynthesis.BMC Plant Biol. 2018 Nov 8;18(1):272. doi: 10.1186/s12870-018-1505-0. BMC Plant Biol. 2018. PMID: 30409115 Free PMC article.
-
Biochemical and Transcriptome Analysis Reveals Pigment Biosynthesis Influenced Chlorina Leaf Formation in Anoectochilus roxburghii (Wall.) Lindl.Biochem Genet. 2024 Apr;62(2):1040-1054. doi: 10.1007/s10528-023-10432-7. Epub 2023 Aug 1. Biochem Genet. 2024. PMID: 37528284
-
Full-Length Transcriptome Analysis Reveals Candidate Genes Involved in Terpenoid Biosynthesis in Artemisia argyi.Front Genet. 2021 Jun 22;12:659962. doi: 10.3389/fgene.2021.659962. eCollection 2021. Front Genet. 2021. PMID: 34239538 Free PMC article.
-
Isolation, structural and bioactivities of polysaccharides from Anoectochilus roxburghii (Wall.) Lindl.: A review.Int J Biol Macromol. 2023 May 1;236:123883. doi: 10.1016/j.ijbiomac.2023.123883. Epub 2023 Mar 6. Int J Biol Macromol. 2023. PMID: 36889614 Review.
-
Recent progress and new perspectives for diterpenoid biosynthesis in medicinal plants.Med Res Rev. 2021 Nov;41(6):2971-2997. doi: 10.1002/med.21816. Epub 2021 May 2. Med Res Rev. 2021. PMID: 33938025 Review.
Cited by
-
Study on the Mechanism of Formononetin Against Hepatocellular Carcinoma: Regulating Metabolic Pathways of Ferroptosis and Cell Cycle.Int J Mol Sci. 2025 Mar 13;26(6):2578. doi: 10.3390/ijms26062578. Int J Mol Sci. 2025. PMID: 40141219 Free PMC article.
-
Comprehensive characterization of volatile terpenoids and terpene synthases in Lanxangia tsaoko.Mol Hortic. 2025 Apr 3;5(1):20. doi: 10.1186/s43897-024-00140-0. Mol Hortic. 2025. PMID: 40176169 Free PMC article.
-
Integrated Transcriptomic and Metabolomic Analysis Reveals Tissue-Specific Flavonoid Biosynthesis and MYB-Mediated Regulation of UGT71A1 in Panax quinquefolius.Int J Mol Sci. 2025 Mar 16;26(6):2669. doi: 10.3390/ijms26062669. Int J Mol Sci. 2025. PMID: 40141311 Free PMC article.
-
Functional Identification and Transcriptional Activity Analysis of Dryopteris fragrans HMGR Gene.Plants (Basel). 2025 Jul 15;14(14):2190. doi: 10.3390/plants14142190. Plants (Basel). 2025. PMID: 40733426 Free PMC article.
-
Integrated metabolomics and transcriptomics analysis comprehensively revealed the underlying metabolite mechanism difference between Male and Female Eucommia ulmoides Oliver.BMC Plant Biol. 2025 May 2;25(1):573. doi: 10.1186/s12870-025-06575-x. BMC Plant Biol. 2025. PMID: 40316915 Free PMC article.
References
LinkOut - more resources
Full Text Sources

