Actin cytoskeleton depolymerization increases matrix metalloproteinase gene expression in breast cancer cells by promoting translocation of cysteine-rich protein 2 to the nucleus
- PMID: 37266453
- PMCID: PMC10229898
- DOI: 10.3389/fcell.2023.1100938
Actin cytoskeleton depolymerization increases matrix metalloproteinase gene expression in breast cancer cells by promoting translocation of cysteine-rich protein 2 to the nucleus
Abstract
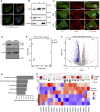
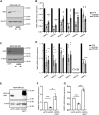
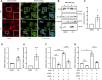
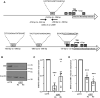
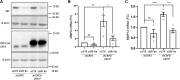
The actin cytoskeleton plays a critical role in cancer cell invasion and metastasis; however, the coordination of its multiple functions remains unclear. Actin dynamics in the cytoplasm control the formation of invadopodia, which are membrane protrusions that facilitate cancer cell invasion by focusing the secretion of extracellular matrix-degrading enzymes, including matrix metalloproteinases (MMPs). In this study, we investigated the nuclear role of cysteine-rich protein 2 (CRP2), a two LIM domain-containing F-actin-binding protein that we previously identified as a cytoskeletal component of invadopodia, in breast cancer cells. We found that F-actin depolymerization stimulates the translocation of CRP2 into the nucleus, resulting in an increase in the transcript levels of pro-invasive and pro-metastatic genes, including several members of the MMP gene family. We demonstrate that in the nucleus, CRP2 interacts with the transcription factor serum response factor (SRF), which is crucial for the expression of MMP-9 and MMP-13. Our data suggest that CRP2 and SRF cooperate to modulate of MMP expression levels. Furthermore, Kaplan-Meier analysis revealed a significant association between high-level expression of SRF and shorter overall survival and distant metastasis-free survival in breast cancer patients with a high CRP2 expression profile. Our findings suggest a model in which CRP2 mediates the coordination of cytoplasmic and nuclear processes driven by actin dynamics, ultimately resulting in the induction of invasive and metastatic behavior in breast cancer cells.
Keywords: Actin cytoskeleton; breast cancer; cysteine-rich protein 2 (CRP2); gene transcription; matrix metalloproteinases (MMPs); metastasis; serum response factor (SRF).
Copyright © 2023 Mgrditchian, Brown-Clay, Hoffmann, Müller, Filali, Ockfen, Mao, Moreau, Casellas, Kaoma, Mittelbronn and Thomas.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

