A plausible identifiable model of the canonical NF-κB signaling pathway
- PMID: 37267242
- PMCID: PMC10237389
- DOI: 10.1371/journal.pone.0286416
A plausible identifiable model of the canonical NF-κB signaling pathway
Abstract
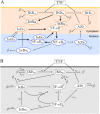
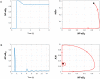
An overwhelming majority of mathematical models of regulatory pathways, including the intensively studied NF-κB pathway, remains non-identifiable, meaning that their parameters may not be determined by existing data. The existing NF-κB models that are capable of reproducing experimental data contain non-identifiable parameters, whereas simplified models with a smaller number of parameters exhibit dynamics that differs from that observed in experiments. Here, we reduced an existing model of the canonical NF-κB pathway by decreasing the number of equations from 15 to 6. The reduced model retains two negative feedback loops mediated by IκBα and A20, and in response to both tonic and pulsatile TNF stimulation exhibits dynamics that closely follow that of the original model. We carried out the sensitivity-based linear analysis and Monte Carlo-based analysis to demonstrate that the resulting model is both structurally and practically identifiable given measurements of 5 model variables from a simple TNF stimulation protocol. The reduced model is capable of reproducing different types of responses that are characteristic to regulatory motifs controlled by negative feedback loops: nearly-perfect adaptation as well as damped and sustained oscillations. It can serve as a building block of more comprehensive models of the immune response and cancer, where NF-κB plays a decisive role. Our approach, although may not be automatically generalized, suggests that models of other regulatory pathways can be transformed to identifiable, while retaining their dynamical features.
Copyright: © 2023 Jaruszewicz-Błońska et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- Wieland F-G, Hauber AL, Rosenblatt M, Tönsing C, Timmer J. On structural and practical identifiability. Curr Opin Syst Biol. 2021; 25:60–9. doi: 10.1016/j.coisb.2021.03.005 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

