Immune engineered extracellular vesicles to modulate T cell activation in the context of type 1 diabetes
- PMID: 37267353
- PMCID: PMC10765990
- DOI: 10.1126/sciadv.adg1082
Immune engineered extracellular vesicles to modulate T cell activation in the context of type 1 diabetes
Abstract
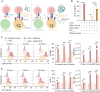
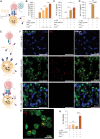
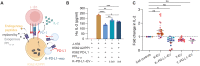
Extracellular vesicles (EVs) can affect immune responses through antigen presentation and costimulation or coinhibition. We generated designer EVs to modulate T cells in the context of type 1 diabetes, a T cell-mediated autoimmune disease, by engineering a lymphoblast cell line, K562, to express HLA-A*02 (HLA-A2) alongside costimulatory CD80 and/or coinhibitory programmed death ligand 1 (PD-L1). EVs presenting HLA-A2 and CD80 activated CD8+ T cells in a dose, antigen, and HLA-specific manner. Adding PD-L1 to these EVs produced an immunoregulatory response, reducing CD8+ T cell activation and cytotoxicity in vitro. EVs alone could not stimulate T cells without antigen-presenting cells. EVs lacking CD80 were ineffective at modulating CD8+ T cell activation, suggesting that both peptide-HLA complex and costimulation are required for EV-mediated immune modulation. These results provide mechanistic insight into the rational design of EVs as a cell-free approach to immunotherapy that can be tailored to promote inflammatory or tolerogenic immune responses.
Figures




References
-
- van Niel G., D'Angelo G., Raposo G., Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228 (2018). - PubMed
-
- Thery C., Witwer K. W., Aikawa E., Alcaraz M. J., Anderson J. D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G. K., Ayre D. C., Bach J. M., Bachurski D., Baharvand H., Balaj L., Baldacchino S., Bauer N. N., Baxter A. A., Bebawy M., Beckham C., Zavec A. B., Benmoussa A., Berardi A. C., Bergese P., Bielska E., Blenkiron C., Bobis-Wozowicz S., Boilard E., Boireau W., Bongiovanni A., Borras F. E., Bosch S., Boulanger C. M., Breakefield X., Breglio A. M., Brennan M. A., Brigstock D. R., Brisson A., Broekman M. L., Bromberg J. F., Bryl-Gorecka P., Buch S., Buck A. H., Burger D., Busatto S., Buschmann D., Bussolati B., Buzas E. I., Byrd J. B., Camussi G., Carter D. R., Caruso S., Chamley L. W., Chang Y. T., Chen C., Chen S., Cheng L., Chin A. R., Clayton A., Clerici S. P., Cocks A., Cocucci E., Coffey R. J., Cordeiro-da-Silva A., Couch Y., Coumans F. A., Coyle B., Crescitelli R., Criado M. F., D'Souza-Schorey C., Das S., Chaudhuri A. D., de Candia P., De Santana E. F., De Wever O., Del Portillo H. A., Demaret T., Deville S., Devitt A., Dhondt B., Di Vizio D., Dieterich L. C., Dolo V., Rubio A. P. D., Dominici M., Dourado M. R., Driedonks T. A., Duarte F. V., Duncan H. M., Eichenberger R. M., Ekstrom K., El Andaloussi S., Elie-Caille C., Erdbrugger U., Falcon-Perez J. M., Fatima F., Fish J. E., Flores-Bellver M., Forsonits A., Frelet-Barrand A., Fricke F., Fuhrmann G., Gabrielsson S., Gamez-Valero A., Gardiner C., Gartner K., Gaudin R., Gho Y. S., Giebel B., Gilbert C., Gimona M., Giusti I., Goberdhan D. C., Gorgens A., Gorski S. M., Greening D. W., Gross J. C., Gualerzi A., Gupta G. N., Gustafson D., Handberg A., Haraszti R. A., Harrison P., Hegyesi H., Hendrix A., Hill A. F., Hochberg F. H., Hoffmann K. F., Holder B., Holthofer H., Hosseinkhani B., Hu G., Huang Y., Huber V., Hunt S., Ibrahim A. G., Ikezu T., Inal J. M., Isin M., Ivanova A., Jackson H. K., Jacobsen S., Jay S. M., Jayachandran M., Jenster G., Jiang L., Johnson S. M., Jones J. C., Jong A., Jovanovic-Talisman T., Jung S., Kalluri R., Kano S. I., Kaur S., Kawamura Y., Keller E. T., Khamari D., Khomyakova E., Khvorova A., Kierulf P., Kim K. P., Kislinger T., Klingeborn M., Klinke D. J. II, Kornek M., Kosanovic M. M., Kovacs A. F., Kramer-Albers E. M., Krasemann S., Krause M., Kurochkin I. V., Kusuma G. D., Kuypers S., Laitinen S., Langevin S. M., Languino L. R., Lannigan J., Lasser C., Laurent L. C., Lavieu G., Lazaro-Ibanez E., Le Lay S., Lee M. S., Lee Y. X. F., Lemos D. S., Lenassi M., Leszczynska A., Li I. T., Liao K., Libregts S. F., Ligeti E., Lim R., Lim S. K., Line A., Linnemannstons K., Llorente A., Lombard C. A., Lorenowicz M. J., Lorincz A. M., Lotvall J., Lovett J., Lowry M. C., Loyer X., Lu Q., Lukomska B., Lunavat T. R., Maas S. L., Malhi H., Marcilla A., Mariani J., Mariscal J., Martens-Uzunova E. S., Martin-Jaular L., Martinez M. C., Martins V. R., Mathieu M., Mathivanan S., Maugeri M., McGinnis L. K., McVey M. J., Meckes D. G. Jr., Meehan K. L., Mertens I., Minciacchi V. R., Moller A., Jorgensen M. M., Morales-Kastresana A., Morhayim J., Mullier F., Muraca M., Musante L., Mussack V., Muth D. C., Myburgh K. H., Najrana T., Nawaz M., Nazarenko I., Nejsum P., Neri C., Neri T., Nieuwland R., Nimrichter L., Nolan J. P., Nolte-'t Hoen E. N., Hooten N. N., O'Driscoll L., O'Grady T., O'Loghlen A., Ochiya T., Olivier M., Ortiz A., Ortiz L. A., Osteikoetxea X., Ostergaard O., Ostrowski M., Park J., Pegtel D. M., Peinado H., Perut F., Pfaffl M. W., Phinney D. G., Pieters B. C., Pink R. C., Pisetsky D. S., von Strandmann E. P., Polakovicova I., Poon I. K., Powell B. H., Prada I., Pulliam L., Quesenberry P., Radeghieri A., Raffai R. L., Raimondo S., Rak J., Ramirez M. I., Raposo G., Rayyan M. S., Regev-Rudzki N., Ricklefs F. L., Robbins P. D., Roberts D. D., Rodrigues S. C., Rohde E., Rome S., Rouschop K. M., Rughetti A., Russell A. E., Saa P., Sahoo S., Salas-Huenuleo E., Sanchez C., Saugstad J. A., Saul M. J., Schiffelers R. M., Schneider R., Schoyen T. H., Scott A., Shahaj E., Sharma S., Shatnyeva O., Shekari F., Shelke G. V., Shetty A. K., Shiba K., Siljander P. R., Silva A. M., Skowronek A., Snyder O. L. II, Soares R. P., Sodar B. W., Soekmadji C., Sotillo J., Stahl P. D., Stoorvogel W., Stott S. L., Strasser E. F., Swift S., Tahara H., Tewari M., Timms K., Tiwari S., Tixeira R., Tkach M., Toh W. S., Tomasini R., Torrecilhas A. C., Tosar J. P., Toxavidis V., Urbanelli L., Vader P., van Balkom B. W., van der Grein S. G., Van Deun J., van Herwijnen M. J., Van Keuren-Jensen K., van Niel G., van Royen M. E., van Wijnen A. J., Vasconcelos M. H., Vechetti I. J. Jr., Veit T. D., Vella L. J., Velot E., Verweij F. J., Vestad B., Vinas J. L., Visnovitz T., Vukman K. V., Wahlgren J., Watson D. C., Wauben M. H., Weaver A., Webber J. P., Weber V., Wehman A. M., Weiss D. J., Welsh J. A., Wendt S., Wheelock A. M., Wiener Z., Witte L., Wolfram J., Xagorari A., Xander P., Xu J., Yan X., Yanez-Mo M., Yin H., Yuana Y., Zappulli V., Zarubova J., Zekas V., Zhang J. Y., Zhao Z., Zheng L., Zheutlin A. R., Zickler A. M., Zimmermann P., Zivkovic A. M., Zocco D., Zuba-Surma E. K., Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell Vesicles 7, 1535750 (2018). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

