Predicting benefit from immune checkpoint inhibitors in patients with non-small-cell lung cancer by CT-based ensemble deep learning: a retrospective study
- PMID: 37268451
- PMCID: PMC10330920
- DOI: 10.1016/S2589-7500(23)00082-1
Predicting benefit from immune checkpoint inhibitors in patients with non-small-cell lung cancer by CT-based ensemble deep learning: a retrospective study
Erratum in
-
Correction to Lancet Digit Health 2023; 5: e404-20.Lancet Digit Health. 2023 Jul;5(7):e403. doi: 10.1016/S2589-7500(23)00110-3. Lancet Digit Health. 2023. PMID: 37391264 Free PMC article. No abstract available.
Abstract
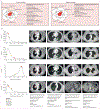
Background: Only around 20-30% of patients with non-small-cell lung cancer (NCSLC) have durable benefit from immune-checkpoint inhibitors. Although tissue-based biomarkers (eg, PD-L1) are limited by suboptimal performance, tissue availability, and tumour heterogeneity, radiographic images might holistically capture the underlying cancer biology. We aimed to investigate the application of deep learning on chest CT scans to derive an imaging signature of response to immune checkpoint inhibitors and evaluate its added value in the clinical context.
Methods: In this retrospective modelling study, 976 patients with metastatic, EGFR/ALK negative NSCLC treated with immune checkpoint inhibitors at MD Anderson and Stanford were enrolled from Jan 1, 2014, to Feb 29, 2020. We built and tested an ensemble deep learning model on pretreatment CTs (Deep-CT) to predict overall survival and progression-free survival after treatment with immune checkpoint inhibitors. We also evaluated the added predictive value of the Deep-CT model in the context of existing clinicopathological and radiological metrics.
Findings: Our Deep-CT model demonstrated robust stratification of patient survival of the MD Anderson testing set, which was validated in the external Stanford set. The performance of the Deep-CT model remained significant on subgroup analyses stratified by PD-L1, histology, age, sex, and race. In univariate analysis, Deep-CT outperformed the conventional risk factors, including histology, smoking status, and PD-L1 expression, and remained an independent predictor after multivariate adjustment. Integrating the Deep-CT model with conventional risk factors demonstrated significantly improved prediction performance, with overall survival C-index increases from 0·70 (clinical model) to 0·75 (composite model) during testing. On the other hand, the deep learning risk scores correlated with some radiomics features, but radiomics alone could not reach the performance level of deep learning, indicating that the deep learning model effectively captured additional imaging patterns beyond known radiomics features.
Interpretation: This proof-of-concept study shows that automated profiling of radiographic scans through deep learning can provide orthogonal information independent of existing clinicopathological biomarkers, bringing the goal of precision immunotherapy for patients with NSCLC closer.
Funding: National Institutes of Health, Mark Foundation Damon Runyon Foundation Physician Scientist Award, MD Anderson Strategic Initiative Development Program, MD Anderson Lung Moon Shot Program, Andrea Mugnaini, and Edward L C Smith.
Copyright © 2023 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY-NC-ND 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests NIV receives consulting fees from Sanofi, Regeneron, Oncocyte, and Eli Lilly; and research funding from Mirati, outside the submitted work. SHL reports research funding from STCube Pharmaceuticals, Beyond Spring Pharmaceuticals, and Nektar Therapeutics; being on an advisory board for AstraZeneca and Creatv Microtech; and receiving consultation fees from XRAD Therapeutics, all outside the submitted work. PPL reports personal fees from Viewray and AstraZeneca; personal fees and non-financial support from Varian; and personal fees from Genentech, outside the submitted work. SG reports research support from AstraZeneca, BMS, and Millenium Pharmaceuticals, all outside the submitted work. JYC reports research funding from BMS-MDACC and consultation fees from Legion Healthcare Partners. MFG reports research support from Varian Medical Systems and RefleXion Medical. HAW reports research funding from ACEA Biosciences, Arrys Therapeutics, AstraZeneca/Medimmune, BMS, Clovis Oncology, Genentech/Roche, Merck, Novartis, SeaGen, Xcovery, and Helsinn; being on an advisory board for AstraZeneca, Blueprint, Mirati, Merck, and Genentech/Roche; and has leadership roles with the International Association for the Study of Lung Cancer, and ECOG-ACRIN. JWN reports honoraria from CME Matters, Clinical Care Options Continuing Medical Education (CME), Research to Practice CME, Medscape CME, Biomedical Learning Institute CME, MLI Peerview CME, Prime Oncology CME, Projects in Knowledge CME, Rockpointe CME, MJH Life Sciences CME, Medical Educator Consortium, and HMP Education; consulting or advisory roles for AstraZeneca, Genentech/Roche, Exelixis, Jounce Therapeutics, Takeda Pharmaceuticals, Eli Lilly, Calithera Biosciences, Amgen, Iovance Biotherapeutics, Blueprint Pharmaceuticals, Regeneron Pharmaceuticals, Natera, Sanofi/Regeneron, D2G Oncology, Surface Oncology, Turning Point Therapeutics, Mirati Therapeutics, Gilead Sciences, and AbbVie; and research funding from Genentech/Roche, Merck, Novartis, Boehringer Ingelheim, Exelixis, Nektar Therapeutics, Takeda Pharmaceuticals, Adaptimmune, GSK, Janssen, and AbbVie. H-SL reports research funding from Samyang Biopharmaceutical USA. VV reports consulting fees from BMS, Merck, Novartis, Amgen, Foundation Medicine, and AstraZeneca. YL reports research funding from Merck, MacroGenics, Tolero Pharmaceuticals, AstraZeneca, Vaccinex, Blueprint Medicines, Harpoon Therapeutics, Sun Pharma Advanced Research, Bristol Myers Squibb, Kyowa Pharmaceuticals, Tesaro, Bayer HealthCare, Mirati Therapeutics, and Daiichi Sankyo; has been on scientific advisory boards for AstraZeneca Pharmaceuticals, Janssen Pharmaceutical, Lilly Oncology, and Turning Point Therapeutics; has received consultation fees from AstraZeneca; and has received honoraria from Clarion Health Care. MP reports research funding from Novartis Institutes for Biomedical Research. XLe reports research funding from Eli Lilly, EMD Serono, Regeneron, and Boehringer Ingelheim; and consultant fees from EMD Serono (Merck KGaA), AstraZeneca, Spectrum Pharmaceutics, Novartis, Eli Lilly, Boehringer Ingelheim, Hengrui Therapeutics, Janssen, Blueprint Medicines, Sensei Biotherapeutics, and AbbVie, outside the submitted work. YYE discloses research support from AstraZeneca, Takeda, Eli Lilly, Xcovery, Tuning Point Therapeutics, Blueprint, Elevation Oncology, Spectrum, and Nuvalent; having advisory roles for AstraZeneca, Eli Lilly, Takeda, Specturm, Bristol Myers Squibb, and Turning Point Therapeutics; and accommodation expenses from Eli Lilly. MVN has been on scientific advisory boards for Mirati, Merck/MSD, and Genentech; and has received research funding from Mirati, Novartis, Checkmate, Alaunos/Ziopharm, AstraZeneca, Pfizer, and Genentech. FS reports consulting fees and advisory roles from Amgen, AstraZeneca Pharmaceuticals, Novartis, BeiGene, Tango Therapeutics, Calithera Biosciences, Navire Pharma, Medscape, Intellisphere, Guardant Health, and BergenBio; speaker fees from BMS, RV Mais Promoção e Eventos, Visiting Speakers Programme in Oncology at McGill University and the Université de Montréal, AIM Group International, and ESMO; fees for travel, food, and beverages from Tango Therapeutics, AstraZeneca Pharmaceuticals, Amgen, Guardant Health, and Dava Oncology; stock or stock options in BioNTech and Moderna; research grants (to institution) from Amgen, Mirati Therapeutics, Boehringer Ingelheim, Merck & Co, and Novartis; study chair funds (to institution) from Pfizer; and research grants (spouse, to institution) from Almmune. CMG reports fees for advisory committees from AstraZeneca, Bristol Myers Squibb, Jazz Pharmaceuticals, and Monte Rosa Therapeutics; research support from AstraZeneca; and speaker's fees from AstraZeneca and Beigene. TC reports speaker fees and honoraria from The Society for Immunotherapy of Cancer, Bristol Myers Squibb, Roche, Medscape, and PeerView; having an advisory role or receiving consulting fees from AstraZeneca, Bristol Myers Squibb, EMD Serono, Merck & Co, Genentech, and Arrowhead Pharmaceuticals; and institutional research funding from AstraZeneca, Bristol Myers Squibb, and EMD Serono. IIW reports grants and personal fees from Genentech/Roche, Bayer, Bristol Myers Squibb, AstraZeneca, Pfizer, HTG Molecular, Merck, Guardant Health, Novartis, and Amgen; personal fees from GSK, Flame, Sanofi, Daiichi Sankyo, Oncocyte, Janssen, MSD, and Platform Health; and grants from Adaptimmune, Adaptive, 4D, EMD Serono, Takeda, Karus, Iovance, Johnson & Johnson, and Akoya outside the submitted work. JDH is on the Scientific Advisory Board of Imagion Biosystems. DLG reports honoraria for scientific advisory boards from AstraZeneca, Sanofi, Alethia Biotherapeutics, Menarini, Eli Lilly, 4D Pharma, and Onconova; and research support from Janssen, Takeda, Astellas, Ribon Therapeutics, NGM Biopharmaceuticals, Boehringer Ingelheim, Mirati Therapeutics, and AstraZeneca. JVH reports being on scientific advisory boards for AstraZeneca, Boehringer Ingelheim, Genentech, GlaxoSmithKline, Eli Lilly, Novartis, Spectrum, EMD Serono, Sanofi, Takeda, Mirati Therapeutics, BMS, and Janssen Global Services; receiving research support from AstraZeneca, Takeda, Boehringer Ingelheim, and Spectrum; and receiving licensing fees from Spectrum. JZ reports research funding from Merck, Johnson & Johnson, and Novartis; and consultant fees from BMS, Johnson & Johnson, AstraZeneca, Geneplus, OrigMed, Novartis, and Innovent, outside the submitted work. CCW reports research support from Medical Imaging and Data Resource Center from NIBIB/University of Chicago and royalties from Elsevier. All other authors declare no competing interests.
Figures






Comment in
-
Deep learning-based radiomics: pacing immunotherapy in lung cancer.Lancet Digit Health. 2023 Jul;5(7):e396-e397. doi: 10.1016/S2589-7500(23)00086-9. Epub 2023 May 31. Lancet Digit Health. 2023. PMID: 37268450 No abstract available.
References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin 2022; 72: 7–33. - PubMed
-
- Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol 2019; 16: 341–55. - PubMed
-
- Doroshow DB, Bhalla S, Beasley MB, et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol 2021; 18: 345–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

