Lipid droplets are a metabolic vulnerability in melanoma
- PMID: 37268606
- PMCID: PMC10238408
- DOI: 10.1038/s41467-023-38831-9
Lipid droplets are a metabolic vulnerability in melanoma
Abstract
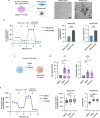
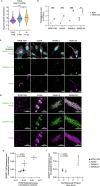
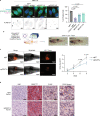
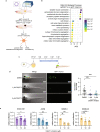
Melanoma exhibits numerous transcriptional cell states including neural crest-like cells as well as pigmented melanocytic cells. How these different cell states relate to distinct tumorigenic phenotypes remains unclear. Here, we use a zebrafish melanoma model to identify a transcriptional program linking the melanocytic cell state to a dependence on lipid droplets, the specialized organelle responsible for lipid storage. Single-cell RNA-sequencing of these tumors show a concordance between genes regulating pigmentation and those involved in lipid and oxidative metabolism. This state is conserved across human melanoma cell lines and patient tumors. This melanocytic state demonstrates increased fatty acid uptake, an increased number of lipid droplets, and dependence upon fatty acid oxidative metabolism. Genetic and pharmacologic suppression of lipid droplet production is sufficient to disrupt cell cycle progression and slow melanoma growth in vivo. Because the melanocytic cell state is linked to poor outcomes in patients, these data indicate a metabolic vulnerability in melanoma that depends on the lipid droplet organelle.
© 2023. The Author(s).
Conflict of interest statement
R.M.W. is a paid consultant to N-of-One Therapeutics, a subsidiary of Qiagen. R.M.W. receives royalty payments for the use of the casper line from Carolina Biologicals. L.S. is co-founder and consultant of BlueRockTherapeutics and is listed as an inventor on a patent application by Memorial Sloan Kettering Cancer Center related to melanocyte differentiation from human pluripotent stem cells (WO2011149762A2). D.L., E.M., E.J., A.B., Y.M., C.L., T.H., and S.S. declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

