Refining the evolutionary tree of the horse Y chromosome
- PMID: 37268661
- PMCID: PMC10238413
- DOI: 10.1038/s41598-023-35539-0
Refining the evolutionary tree of the horse Y chromosome
Abstract
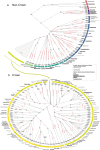
The Y chromosome carries information about the demography of paternal lineages, and thus, can prove invaluable for retracing both the evolutionary trajectory of wild animals and the breeding history of domesticates. In horses, the Y chromosome shows a limited, but highly informative, sequence diversity, supporting the increasing breeding influence of Oriental lineages during the last 1500 years. Here, we augment the primary horse Y-phylogeny, which is currently mainly based on modern horse breeds of economic interest, with haplotypes (HT) segregating in remote horse populations around the world. We analyze target enriched sequencing data of 5 Mb of the Y chromosome from 76 domestic males, together with 89 whole genome sequenced domestic males and five Przewalski's horses from previous studies. The resulting phylogeny comprises 153 HTs defined by 2966 variants and offers unprecedented resolution into the history of horse paternal lineages. It reveals the presence of a remarkable number of previously unknown haplogroups in Mongolian horses and insular populations. Phylogenetic placement of HTs retrieved from 163 archaeological specimens further indicates that most of the present-day Y-chromosomal variation evolved after the domestication process that started around 4200 years ago in the Western Eurasian steppes. Our comprehensive phylogeny significantly reduces ascertainment bias and constitutes a robust evolutionary framework for analyzing horse population dynamics and diversity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

