NFYA promotes malignant behavior of triple-negative breast cancer in mice through the regulation of lipid metabolism
- PMID: 37268670
- PMCID: PMC10238388
- DOI: 10.1038/s42003-023-04987-9
NFYA promotes malignant behavior of triple-negative breast cancer in mice through the regulation of lipid metabolism
Abstract
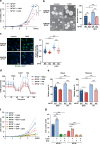
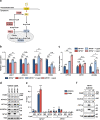
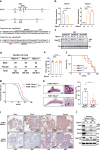
Two splicing variants exist in NFYA that exhibit high expression in many human tumour types. The balance in their expression correlates with prognosis in breast cancer, but functional differences remain unclear. Here, we demonstrate that NFYAv1, a long-form variant, upregulates the transcription of essential lipogenic enzymes ACACA and FASN to enhance the malignant behavior of triple-negative breast cancer (TNBC). Loss of the NFYAv1-lipogenesis axis strongly suppresses malignant behavior in vitro and in vivo, indicating that the NFYAv1-lipogenesis axis is essential for TNBC malignant behavior and that the axis might be a potential therapeutic target for TNBC. Furthermore, mice deficient in lipogenic enzymes, such as Acly, Acaca, and Fasn, exhibit embryonic lethality; however, Nfyav1-deficient mice exhibited no apparent developmental abnormalities. Our results indicate that the NFYAv1-lipogenesis axis has tumour-promoting effects and that NFYAv1 may be a safe therapeutic target for TNBC.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

