Increase in rotavirus prevalence with the emergence of genotype G9P[8] in replacement of genotype G12P[6] in Sabah, Malaysia
- PMID: 37269384
- PMCID: PMC10238249
- DOI: 10.1007/s00705-023-05803-9
Increase in rotavirus prevalence with the emergence of genotype G9P[8] in replacement of genotype G12P[6] in Sabah, Malaysia
Abstract
Rotaviruses are major causative agents of acute diarrhea in children under 5 years of age in Malaysia. However, a rotavirus vaccine has not been included in the national vaccination program. To date, only two studies have been carried out in the state of Sabah, Malaysia, although children in this state are at risk of diarrheal diseases. Previous studies showed that 16%-17% of cases of diarrhea were caused by rotaviruses and that equine-like G3 rotavirus strains are predominant. Because the prevalence of rotaviruses and their genotype distribution vary over time, this study was conducted at four government healthcare facilities from September 2019 through February 2020. Our study revealed that the proportion of rotavirus diarrhea increased significantly to 37.2% (51/137) after the emergence of the G9P[8] genotype in replacement of the G12P[8] genotype. Although equine-like G3P[8] strains remain the predominant rotaviruses circulating among children, the Sabahan G9P[8] strain belonged to lineage VI and was phylogenetically related to strains from other countries. A comparison of the Sabahan G9 strains with the G9 vaccine strains used in the RotaSiil and Rotavac vaccines revealed several mismatches in neutralizing epitopes, indicating that these vaccines might not be effective in Sabahan children. However, a vaccine trial may be necessary to understand the precise effects of vaccination.
© 2023. The Author(s), under exclusive licence to Springer-Verlag GmbH Austria, part of Springer Nature.
Conflict of interest statement
All authors declare that they have no conflict of interest.
Figures
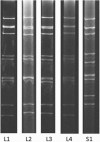






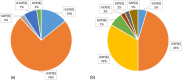
Similar articles
-
Emergence of equine-like G3 strains as the dominant rotavirus among children under five with diarrhea in Sabah, Malaysia during 2018-2019.PLoS One. 2021 Jul 28;16(7):e0254784. doi: 10.1371/journal.pone.0254784. eCollection 2021. PLoS One. 2021. PMID: 34320003 Free PMC article.
-
Epidemiology and genetic diversity of group A rotavirus in acute diarrhea patients in pre-vaccination era in Himachal Pradesh, India.Vaccine. 2019 Aug 23;37(36):5350-5356. doi: 10.1016/j.vaccine.2019.07.037. Epub 2019 Jul 19. Vaccine. 2019. PMID: 31331769
-
Emergence of equine-like G3 and porcine-like G9 rotavirus strains in Sarawak, Malaysia: 2019-2021.J Med Virol. 2023 Aug;95(8):e28987. doi: 10.1002/jmv.28987. J Med Virol. 2023. PMID: 37501648
-
A decade of G3P[8] and G9P[8] rotaviruses in Brazil: epidemiology and evolutionary analyses.Infect Genet Evol. 2014 Dec;28:389-97. doi: 10.1016/j.meegid.2014.05.016. Epub 2014 May 23. Infect Genet Evol. 2014. PMID: 24861814
-
Driving forces of continuing evolution of rotaviruses.World J Virol. 2024 Jun 25;13(2):93774. doi: 10.5501/wjv.v13.i2.93774. World J Virol. 2024. PMID: 38984077 Free PMC article. Review.
Cited by
-
Comparative analysis of the RVA VP7 and VP4 antigenic epitopes circulating in Iran and the Rotarix and RotaTeq vaccines.Heliyon. 2024 Jul 4;10(13):e33887. doi: 10.1016/j.heliyon.2024.e33887. eCollection 2024 Jul 15. Heliyon. 2024. PMID: 39071626 Free PMC article.
-
Changes in vaccine coverage and incidence of acute gastroenteritis and severe rotavirus gastroenteritis in children <5 years in Shibata City, Niigata Prefecture, Japan.Hum Vaccin Immunother. 2024 Dec 31;20(1):2322202. doi: 10.1080/21645515.2024.2322202. Epub 2024 Mar 13. Hum Vaccin Immunother. 2024. PMID: 38478958 Free PMC article.
-
Outbreak report of rotavirus gastroenteritis among remotely vaccinated travelers: A potential implication of booster vaccine for travelers to endemic countries.Hum Vaccin Immunother. 2025 Dec;21(1):2467475. doi: 10.1080/21645515.2025.2467475. Epub 2025 Feb 26. Hum Vaccin Immunother. 2025. PMID: 40008469 Free PMC article.
-
Temporal changes in the positivity rate of common enteric viruses among paediatric admissions in coastal Kenya, during the COVID-19 pandemic, 2019-2022.Gut Pathog. 2024 Jan 4;16(1):2. doi: 10.1186/s13099-023-00595-4. Gut Pathog. 2024. PMID: 38178245 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

