Quantitative trait and transcriptome analysis of genetic complexity underpinning cardiac interatrial septation in mice using an advanced intercross line
- PMID: 37272612
- PMCID: PMC10284603
- DOI: 10.7554/eLife.83606
Quantitative trait and transcriptome analysis of genetic complexity underpinning cardiac interatrial septation in mice using an advanced intercross line
Abstract
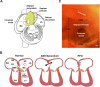
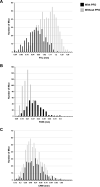
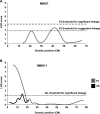
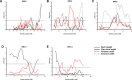
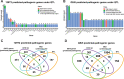
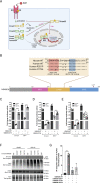
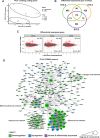
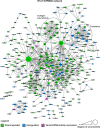
Unlike single-gene mutations leading to Mendelian conditions, common human diseases are likely to be emergent phenomena arising from multilayer, multiscale, and highly interconnected interactions. Atrial and ventricular septal defects are the most common forms of cardiac congenital anomalies in humans. Atrial septal defects (ASD) show an open communication between the left and right atria postnatally, potentially resulting in serious hemodynamic consequences if untreated. A milder form of atrial septal defect, patent foramen ovale (PFO), exists in about one-quarter of the human population, strongly associated with ischaemic stroke and migraine. The anatomic liabilities and genetic and molecular basis of atrial septal defects remain unclear. Here, we advance our previous analysis of atrial septal variation through quantitative trait locus (QTL) mapping of an advanced intercross line (AIL) established between the inbred QSi5 and 129T2/SvEms mouse strains, that show extremes of septal phenotypes. Analysis resolved 37 unique septal QTL with high overlap between QTL for distinct septal traits and PFO as a binary trait. Whole genome sequencing of parental strains and filtering identified predicted functional variants, including in known human congenital heart disease genes. Transcriptome analysis of developing septa revealed downregulation of networks involving ribosome, nucleosome, mitochondrial, and extracellular matrix biosynthesis in the 129T2/SvEms strain, potentially reflecting an essential role for growth and cellular maturation in septal development. Analysis of variant architecture across different gene features, including enhancers and promoters, provided evidence for the involvement of non-coding as well as protein-coding variants. Our study provides the first high-resolution picture of genetic complexity and network liability underlying common congenital heart disease, with relevance to human ASD and PFO.
Keywords: cardiac congenital defects; developmental biology; genetics; genomics; mouse; quantitative trait loci.
© 2023, Moradi Marjaneh, Kirk, Patrick et al.
Conflict of interest statement
MM, EK, RP, DA, DH, GD, PC, VJ, TD, SD, EW, CM, IM, PT, RH No competing interests declared
Figures

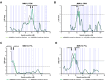
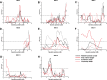
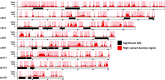
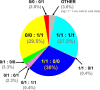

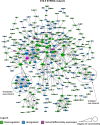


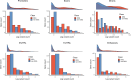

Update of
- doi: 10.1101/2022.10.31.514499
References
-
- Alankarage D, Ip E, Szot JO, Munro J, Blue GM, Harrison K, Cuny H, Enriquez A, Troup M, Humphreys DT, Wilson M, Harvey RP, Sholler GF, Graham RM, Ho JWK, Kirk EP, Pachter N, Chapman G, Winlaw DS, Giannoulatou E, Dunwoodie SL. Identification of clinically actionable variants from genome sequencing of families with congenital heart disease. Genetics in Medicine. 2019;21:1111–1120. doi: 10.1038/s41436-018-0296-x. - DOI - PubMed
-
- Audain E, Wilsdon A, Breckpot J, Izarzugaza JMG, Fitzgerald TW, Kahlert A-K, Sifrim A, Wünnemann F, Perez-Riverol Y, Abdul-Khaliq H, Bak M, Bassett AS, Benson WD, Berger F, Daehnert I, Devriendt K, Dittrich S, Daubeney PE, Garg V, Hackmann K, Hoff K, Hofmann P, Dombrowsky G, Pickardt T, Bauer U, Keavney BD, Klaassen S, Kramer H-H, Marshall CR, Milewicz DM, Lemaire S, Coselli JS, Mitchell ME, Tomita-Mitchell A, Prakash SK, Stamm K, Stewart AFR, Silversides CK, Siebert R, Stiller B, Rosenfeld JA, Vater I, Postma AV, Caliebe A, Brook JD, Andelfinger G, Hurles ME, Thienpont B, Larsen LA, Hitz M-P, Firulli AB. Integrative analysis of genomic variants reveals new associations of candidate haploinsufficient genes with congenital heart disease. PLOS Genetics. 2021;17:e1009679. doi: 10.1371/journal.pgen.1009679. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

