Notch directs telencephalic development and controls neocortical neuron fate determination by regulating microRNA levels
- PMID: 37272771
- PMCID: PMC10309580
- DOI: 10.1242/dev.201408
Notch directs telencephalic development and controls neocortical neuron fate determination by regulating microRNA levels
Abstract
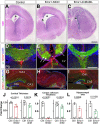
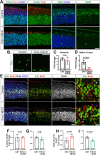
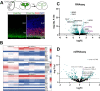
The central nervous system contains a myriad of different cell types produced from multipotent neural progenitors. Neural progenitors acquire distinct cell identities depending on their spatial position, but they are also influenced by temporal cues to give rise to different cell populations over time. For instance, the progenitors of the cerebral neocortex generate different populations of excitatory projection neurons following a well-known sequence. The Notch signaling pathway plays crucial roles during this process, but the molecular mechanisms by which Notch impacts progenitor fate decisions have not been fully resolved. Here, we show that Notch signaling is essential for neocortical and hippocampal morphogenesis, and for the development of the corpus callosum and choroid plexus. Our data also indicate that, in the neocortex, Notch controls projection neuron fate determination through the regulation of two microRNA clusters that include let-7, miR-99a/100 and miR-125b. Our findings collectively suggest that balanced Notch signaling is crucial for telencephalic development and that the interplay between Notch and miRNAs is essential for the control of neocortical progenitor behaviors and neuron cell fate decisions.
Keywords: Cell fate; Cortical development; Mouse; Neurogenesis; Notch; miRNA.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

