Analytical and Clinical Evaluation of a TaqMan Real-Time PCR Assay for the Detection of Chikungunya Virus
- PMID: 37272795
- PMCID: PMC10433969
- DOI: 10.1128/spectrum.00088-23
Analytical and Clinical Evaluation of a TaqMan Real-Time PCR Assay for the Detection of Chikungunya Virus
Abstract
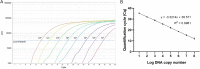
Due to the general symptoms presented by the Chikungunya virus (CHIKV)-infected patients, a laboratory test is needed to differentiate CHIKV from other viral infections. The reverse transcription-quantitative real-time PCR (RT-qPCR) is a rapid and sensitive diagnostic tool, and several assays have been developed for detecting and quantifying CHIKV. Since real-time amplification efficiency varies within and between laboratories, an assay must be validated before being used on patient samples. In this study, the diagnostic performance of a TaqMan RT-qPCR assay was evaluated using synthetic RNA and archived patient samples. The cutoff quantification cycle (Cq) value for the assay was determined by experimental evidence. We found the in-house assay was highly sensitive, with a detection limit of 3.95 RNA copies/reaction. The analytical specificity of the assay was 100%. The analytical cutoff Cq value was 37, corresponding to the mean Cq value of the detection limit. Using archived samples characterized previously, the sensitivity and specificity of the assay were 76% and 100%, respectively. The in-house assay was also compared with a commercial assay, and we found that the in-house assay had higher sensitivity. Although further evaluation with prospective patient samples is needed in the future, this validated RT-qPCR was sensitive and specific, which shows its potential to detect CHIKV in clinical samples. IMPORTANCE Chikungunya virus causes chikungunya fever, a disease characterized by fever, rash, and joint pain. In the early phase of infection, chikungunya fever is always misdiagnosed as other arbovirus infections, such as dengue. Laboratory tests such as RT-qPCR are therefore necessary to confirm CHIKV infection. We evaluated the performance of an in-house RT-qPCR assay, and our study shows that the assay could detect CHIKV in clinical samples. We also show the cutoff determination of the assay, which provides important guidance to scientists or researchers when implementing a new RT-qPCR assay in a laboratory.
Keywords: Chikungunya virus; cutoff quantification cycle; in-house assay; reverse transcription-quantitative real-time PCR.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

