Epigenetic targets in B- and T-cell lymphomas: latest developments
- PMID: 37273421
- PMCID: PMC10236259
- DOI: 10.1177/20406207231173485
Epigenetic targets in B- and T-cell lymphomas: latest developments
Abstract
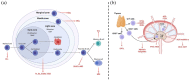
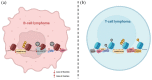
Non-Hodgkin's lymphomas (NHLs) comprise a diverse group of diseases, either of mature B-cell or of T-cell derivation, characterized by heterogeneous molecular features and clinical manifestations. While most of the patients are responsive to standard chemotherapy, immunotherapy, radiation and/or stem cell transplantation, relapsed and/or refractory cases still have a dismal outcome. Deep sequencing analysis have pointed out that epigenetic dysregulations, including mutations in epigenetic enzymes, such as chromatin modifiers and DNA methyltransferases (DNMTs), are prevalent in both B- cell and T-cell lymphomas. Accordingly, over the past decade, a large number of epigenetic-modifying agents have been developed and introduced into the clinical management of these entities, and a few specific inhibitors have already been approved for clinical use. Here we summarize the main epigenetic alterations described in B- and T-NHL, that further supported the clinical development of a selected set of epidrugs in determined diseases, including inhibitors of DNMTs, histone deacetylases (HDACs), and extra-terminal domain proteins (bromodomain and extra-terminal motif; BETs). Finally, we highlight the most promising future directions of research in this area, explaining how bioinformatics approaches can help to identify new epigenetic targets in B- and T-cell lymphoid neoplasms.
Keywords: BET inhibitors; DNMT; EZH2; HAT; HDAC; bioinformatics; clinical testing; drug combination; epigenetics; non-Hodgkin’s lymphoma.
© The Author(s), 2023.
Conflict of interest statement
The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: G. Roué received research funding from TG Therapeutics, Inc. and Kancera AB, to support studies unrelated to this work. The remaining authors have no competing financial interests.
Figures
References
-
- Seifert M, Scholtysik R, Küppers R. Origin and pathogenesis of B cell lymphomas. Methods Mol Biol 2013; 971: 1–25. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

