Genome sequencing with comprehensive variant calling identifies structural variants and repeat expansions in a large fraction of individuals with ataxia and/or neuromuscular disorders
- PMID: 37273706
- PMCID: PMC10234573
- DOI: 10.3389/fneur.2023.1170005
Genome sequencing with comprehensive variant calling identifies structural variants and repeat expansions in a large fraction of individuals with ataxia and/or neuromuscular disorders
Abstract
Introduction: Neuromuscular disorders (NMDs) have a heterogeneous etiology. A genetic diagnosis is key to personalized healthcare and access to targeted treatment for the affected individuals.
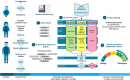
Methods: In this study, 861 patients with NMDs were analyzed with genome sequencing and comprehensive variant calling including single nucleotide variants, small insertions/deletions (SNVs/INDELs), and structural variants (SVs) in a panel of 895 NMD genes, as well as short tandem repeat expansions (STRs) at 28 loci. In addition, for unsolved cases with an unspecific clinical presentation, the analysis of a panel with OMIM disease genes was added.
Results: In the cohort, 27% (232/861) of the patients harbored pathogenic variants, of which STRs and SVs accounted for one-third of the patients (71/232). The variants were found in 107 different NMD genes. Furthermore, 18 pediatric patients harbored pathogenic variants in non-NMD genes.
Discussion: Our results highlight that for children with unspecific hypotonia, a genome-wide analysis rather than a disease-based gene panel should be considered as a diagnostic approach. More importantly, our results clearly show that it is crucial to include STR- and SV-analyses in the diagnostics of patients with neuromuscular disorders.
Keywords: ataxia; genome sequencing; neuromuscular disorders; repeat expansion; single nucleotide variant; structural variant.
Copyright © 2023 Ek, Nilsson, Engvall, Malmgren, Thonberg, Pettersson, Anderlid, Hammarsjö, Helgadottir, Arnardottir, Naess, Nennesmo, Paucar, Hjartarson, Press, Solders, Sejersen, Lindstrand and Kvarnung.
Conflict of interest statement
AL has received honoraria from Illumina Inc., and has been an advisor for Oxford Nanopore Technologies, both unrelated to the content in this article. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Dalil Hamroun GB, Rivier R. Gene Table of Neuromuscular Disorders. (2023). Available from: https://www.musclegenetable.fr (accessed November 2, 2022).
LinkOut - more resources
Full Text Sources

