Exosomal miR-141-3p from PDLSCs Alleviates High Glucose-Induced Senescence of PDLSCs by Activating the KEAP1-NRF2 Signaling Pathway
- PMID: 37274022
- PMCID: PMC10238146
- DOI: 10.1155/2023/7136819
Exosomal miR-141-3p from PDLSCs Alleviates High Glucose-Induced Senescence of PDLSCs by Activating the KEAP1-NRF2 Signaling Pathway
Abstract
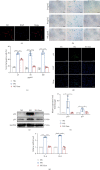
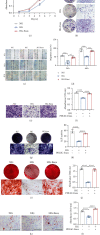
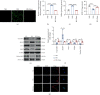
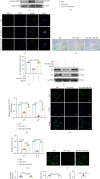
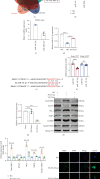
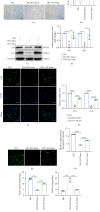
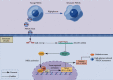
Human periodontal ligament stem cells (PDLSCs) are the most promising stem cells for periodontal tissue engineering. Senescent PDLSCs have diminished abilities to proliferate and differentiate, affecting the efficiency of periodontal tissue repair and regeneration. Stem cell-derived exosomes are important participants in intercellular information exchange and can help ameliorate senescence. In this study, we investigated PDLSC senescence in a high glucose microenvironment as well as the ability of human periodontal ligament stem cell-derived exosomes (PDLSC-Exos) to alleviate cellular senescence and the underlying mechanisms. Herein, PDLSCs and PDLSC-Exos were isolated and extracted. Then, cellular senescence indicators were evaluated after high glucose (25 mM) treatment of cultured PDLSCs. PDLSC-Exos were cocultured with senescent PDLSCs to further explore the role of PDLSC-Exos in cellular senescence and determine the differences in cellular oxidative stress levels after PDLSC-Exo treatment. Next, we investigated whether PDLSC-Exos alleviated cellular senescence by restoring the balance of oxidative stress signals and explored the underlying molecular pathways. We discovered that PDLSCs underwent premature senescence due to high glucose culture, but they were rejuvenated by PDLSC-Exos. The rejuvenating effects of PDLSC-Exos were notably reversed by cotreatment with ML385, an inhibitor of nuclear factor erythroid 2-related factor 2 (NRF2), indicating that this recovery depended on NRF2 activation. Further analyses revealed that microRNA-141-3p (miR-141-3p) was expressed at relatively high levels in PDLSC-Exos and was instrumental in PDLSC-Exo-mediated restoration by downregulating Kelch-like ECH-associated protein 1 (KEAP1), which is a negative regulator of NRF2 expression. Our findings suggest that PDLSC-Exos alleviate high glucose-induced senescence of PDLSCs by transferring miR-141-3p to activate the KEAP1-NRF2 signaling pathway. Based on this research, PDLSC-Exos may behave similarly to their parental PDLSCs and have significant effects on cellular senescence by delivering their encapsulated bioactive chemicals to target cells.
Copyright © 2023 Min Liu et al.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this article.
Figures


Similar articles
-
Human embryonic stem cell-derived exosomes promote pressure ulcer healing in aged mice by rejuvenating senescent endothelial cells.Stem Cell Res Ther. 2019 May 21;10(1):142. doi: 10.1186/s13287-019-1253-6. Stem Cell Res Ther. 2019. PMID: 31113469 Free PMC article.
-
Inflammatory Periodontal Ligament Stem Cells Drive M1 Macrophage Polarization via Exosomal miR-143-3p-Mediated Regulation of PI3K/AKT/NF-κB Signaling.Stem Cells. 2023 Mar 2;41(2):184-199. doi: 10.1093/stmcls/sxac087. Stem Cells. 2023. PMID: 36520505
-
Senolytic elimination of senescent cells improved periodontal ligament stem cell-based bone regeneration partially through inhibiting YAP.Biochim Biophys Acta Mol Cell Res. 2025 Mar;1872(3):119921. doi: 10.1016/j.bbamcr.2025.119921. Epub 2025 Feb 17. Biochim Biophys Acta Mol Cell Res. 2025. PMID: 39971252
-
Periodontal ligament stem cell-based bioactive constructs for bone tissue engineering.Front Bioeng Biotechnol. 2022 Dec 2;10:1071472. doi: 10.3389/fbioe.2022.1071472. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36532583 Free PMC article. Review.
-
The effect of adjunctive LASER application on periodontal ligament stem cells.Front Cell Dev Biol. 2024 Jan 12;11:1341628. doi: 10.3389/fcell.2023.1341628. eCollection 2023. Front Cell Dev Biol. 2024. PMID: 38283989 Free PMC article. Review.
Cited by
-
Nanomaterials Modulating the Fate of Dental-Derived Mesenchymal Stem Cells Involved in Oral Tissue Reconstruction: A Systematic Review.Int J Nanomedicine. 2023 Sep 21;18:5377-5406. doi: 10.2147/IJN.S418675. eCollection 2023. Int J Nanomedicine. 2023. PMID: 37753067 Free PMC article. Review.
-
Diabetes and periodontitis: the role of a high-glucose microenvironment in periodontal tissue cells and corresponding therapeutic strategies.Stem Cell Res Ther. 2025 Jul 15;16(1):366. doi: 10.1186/s13287-025-04441-z. Stem Cell Res Ther. 2025. PMID: 40660319 Free PMC article. Review.
-
Emerging roles of exosomes in oral diseases progression.Int J Oral Sci. 2024 Jan 15;16(1):4. doi: 10.1038/s41368-023-00274-9. Int J Oral Sci. 2024. PMID: 38221571 Free PMC article. Review.
-
The extracellular vesicle-based treatment: a developing strategy for periodontal diseases.Front Immunol. 2025 May 29;16:1480292. doi: 10.3389/fimmu.2025.1480292. eCollection 2025. Front Immunol. 2025. PMID: 40510371 Free PMC article. Review.
-
METTL3 Promotes Osteogenic Differentiation of Human Periodontal Ligament Stem Cells Under the Inflammatory Microenvironment Through the miR-141-3p/ZEB1 Axis.Cell Biochem Biophys. 2025 Jun;83(2):1771-1783. doi: 10.1007/s12013-024-01586-1. Epub 2024 Dec 16. Cell Biochem Biophys. 2025. PMID: 39681812 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

