Drosophila melanogaster as a model to study autophagy in neurodegenerative diseases induced by proteinopathies
- PMID: 37274187
- PMCID: PMC10232775
- DOI: 10.3389/fnins.2023.1082047
Drosophila melanogaster as a model to study autophagy in neurodegenerative diseases induced by proteinopathies
Abstract
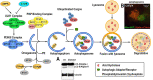
Proteinopathies are a large group of neurodegenerative diseases caused by both genetic and sporadic mutations in particular genes which can lead to alterations of the protein structure and to the formation of aggregates, especially toxic for neurons. Autophagy is a key mechanism for clearing those aggregates and its function has been strongly associated with the ubiquitin-proteasome system (UPS), hence mutations in both pathways have been associated with the onset of neurodegenerative diseases, particularly those induced by protein misfolding and accumulation of aggregates. Many crucial discoveries regarding the molecular and cellular events underlying the role of autophagy in these diseases have come from studies using Drosophila models. Indeed, despite the physiological and morphological differences between the fly and the human brain, most of the biochemical and molecular aspects regulating protein homeostasis, including autophagy, are conserved between the two species.In this review, we will provide an overview of the most common neurodegenerative proteinopathies, which include PolyQ diseases (Huntington's disease, Spinocerebellar ataxia 1, 2, and 3), Amyotrophic Lateral Sclerosis (C9orf72, SOD1, TDP-43, FUS), Alzheimer's disease (APP, Tau) Parkinson's disease (a-syn, parkin and PINK1, LRRK2) and prion diseases, highlighting the studies using Drosophila that have contributed to understanding the conserved mechanisms and elucidating the role of autophagy in these diseases.
Keywords: Drosophila melanogaster; animal model; autophagy; neurodegeneration; non-autonomous signaling; protein-aggregate; protein-misfolding; proteinopathies.
Copyright © 2023 Santarelli, Londero, Soldano, Candelaresi, Todeschini, Vernizzi and Bellosta.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- An H., Skelt L., Notaro A., Highley J. R., Fox A. H., La Bella V., et al. . (2019). ALS-linked FUS mutations confer loss and gain of function in the nucleus by promoting excessive formation of dysfunctional paraspeckles. Acta Neuropathol. Commun. 7:7. doi: 10.1186/s40478-019-0658-x - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

