Genome-wide search identified DNA methylation sites that regulate the metabolome
- PMID: 37274792
- PMCID: PMC10233745
- DOI: 10.3389/fgene.2023.1093882
Genome-wide search identified DNA methylation sites that regulate the metabolome
Abstract
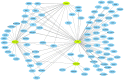
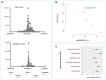
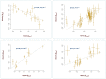
Background: Identifying DNA methylation sites that regulate the metabolome is important for several purposes. In this study, publicly available GWAS data were integrated to find methylation sites that impact metabolome through a discovery and replication scheme and by using Mendelian randomization. Results: The outcome of analyses revealed 107 methylation sites associated with 84 metabolites at the genome-wide significance level (p<5e-8) at both the discovery and replication stages. A large percentage of the observed associations (85%) were with lipids, significantly higher than expected (p = 0.0003). A number of CpG (methylation) sites showed specificity e.g., cg20133200 within PFKP was associated with glucose only and cg10760299 within GATM impacted the level of creatinine; in contrast, there were sites associated with numerous metabolites e.g., cg20102877 on the 2p23.3 region was associated with 39 metabolites. Integrating transcriptome data enabled identifying genes (N = 82) mediating the impact of methylation sites on the metabolome and cardiometabolic traits. For example, PABPC4 mediated the impact of cg15123755-HDL on type-2 diabetes. KCNK7 mediated the impact of cg21033440-lipids on hypertension. POC5, ILRUN, FDFT1, and NEIL2 mediated the impact of CpG sites on obesity through metabolic pathways. Conclusion: This study provides a catalog of DNA methylation sites that regulate the metabolome for downstream applications.
Keywords: DNA methylation; Mendelian randomization; biomarker; epigenomics; metabolimics; metabolite.
Copyright © 2023 Nikpay.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous

