Study of Some Resistance Genes in Clinical Proteus mirabilis
- PMID: 37274906
- PMCID: PMC10237585
- DOI: 10.22092/ARI.2022.358489.2230
Study of Some Resistance Genes in Clinical Proteus mirabilis
Abstract
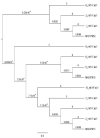
Proteus mirabilis belongs to the family Enterobacteriaceae and is capable of transforming in shape from rod to elongated and swarming motility by flagella. It is an opportunity for bacteria and can cause different clinical diseases. Therefore, this study aimed to assay and detect a sequence of genes that encode for antibiotic resistance in multidrug resistance clinical isolates of Proteus mirabilis, including blaTEM, aac(6')-Ib, qnrA, IntI2, IntI1 and secondly to investigate the relationship in the phylogenetic tree among these genes in Iraq comparison with global strains in NCBI. The study included the identifying of 500 clinical samples depending on morphological and biochemical tests and confirming Proteus mirabilis diagnosis by the VITEK-2 Compact system. The confirmed isolates of Proteus mirabilis were 95 clinical isolates (19%). Antibiotic susceptibility test of all these isolates was done using twelve antibiotics tested using Amoxicillin, Aztreonam, Imipenem, Cefoxitin, Amikacin, Ceftazidem, Ciprofloxacin, Nalidixic acid, Gentamicin, Sulphamethazol-trimethoprim, Cefotaxime, Amoxicillin-clavulanic acid. The results showed that multidrug resistance Proteus mirabilis isolates contained the genes in different levels as follow blaTEM gene (90%), aac(6')-Ib gene (80%) ,IntI1 gene (100%), IntI2 gene (80%). These genes were sequenced and detected phylogenetic relationships among these genes and global genes were documented in NCBI. The results showed that some Iraqi isolates contain genetic variation compared to global strains. Therefore, this variation was detected and registered in NCBI of all five antibiotic resistance genes mentioned above and accepted under accession numbers of aacIb gene (LC613168.1), blaTEM gene (LC613166.1), IntI1 gene (LC613169.1), IntI2 gene (LC613170.1).
Keywords: Antibiotic resistance genes; Integrons; Iraqi clinical samples; Multidrug Proteus mirabilis; NCBI.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







Similar articles
-
Molecular drivers of emerging multidrug resistance in Proteus mirabilis clinical isolates from Algeria.J Glob Antimicrob Resist. 2019 Sep;18:249-256. doi: 10.1016/j.jgar.2019.01.030. Epub 2019 Feb 21. J Glob Antimicrob Resist. 2019. PMID: 30797091
-
First report of blaOXA-24 carbapenemase gene, armA methyltransferase and aac(6')-Ib-cr among multidrug-resistant clinical isolates of Proteus mirabilis in Algeria.J Glob Antimicrob Resist. 2019 Mar;16:125-129. doi: 10.1016/j.jgar.2018.08.019. Epub 2018 Sep 11. J Glob Antimicrob Resist. 2019. PMID: 30217548
-
First report in Africa of two clinical isolates of Proteus mirabilis carrying Salmonella genomic island (SGI1) variants, SGI1-PmABB and SGI1-W.Infect Genet Evol. 2017 Jul;51:132-137. doi: 10.1016/j.meegid.2017.03.029. Epub 2017 Mar 28. Infect Genet Evol. 2017. PMID: 28359833
-
From the Urinary Catheter to the Prevalence of Three Classes of Integrons, β-Lactamase Genes, and Differences in Antimicrobial Susceptibility of Proteus mirabilis and Clonal Relatedness with Rep-PCR.Biomed Res Int. 2021 Jun 10;2021:9952769. doi: 10.1155/2021/9952769. eCollection 2021. Biomed Res Int. 2021. PMID: 34212042 Free PMC article.
-
[Prevalence of Proteus mirabilis strains in clinical specimens and evaluation of their resistance to selected antibiotics].Pol Merkur Lekarski. 2004 Nov;17(101):538-40. Pol Merkur Lekarski. 2004. PMID: 15754653 Review. Polish.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
