Nucleocapsid protein binding DNA aptamers for detection of SARS-COV-2
- PMID: 37275459
- PMCID: PMC10223630
- DOI: 10.1016/j.crbiot.2023.100132
Nucleocapsid protein binding DNA aptamers for detection of SARS-COV-2
Abstract
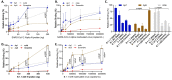
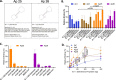
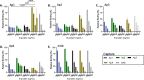
The severe acute respiratory syndrome coronavirus (SARS-CoV-2) has infected millions of individuals and continues to be a major health concern worldwide. While reverse transcription-polymerase chain reaction remains a reliable method for detecting infections, limitations of this technology, particularly cost and the requirement of a dedicated laboratory, prevent rapid viral monitoring. Antigen tests filled this need to some extent but with limitations including sensitivity and specificity, particularly against emerging variants of concern. Here, we developed aptamers against the SARS-CoV-2 Nucleocapsid protein to complement or replace antibodies in antigen detection assays. As detection reagents in ELISA-like assays, our DNA aptamers were able to detect as low as 150 pg/mL of the protein and under 150 k copies of inactivated SARS-CoV-2 Wuhan Alpha strain in viral transport medium with little cross-reactivity to other human coronaviruses (HCoVs). Further, our aptamers were reselected against the SARS-CoV-2 Omicron variant of concern, and we found two sequences that had a more than two-fold increase in signal compared to our original aptamers when used as detection reagents against protein from the Omicron strain. These findings illustrate the use of aptamers as promising alternative detection reagents that may translate for use in current tests and our findings validate the method for the reselection of aptamers against emerging viral strains.
Keywords: Antigen; Aptamer; COVID-19; Diagnostic; Nucleocapsid; SARS-CoV-2.
© 2023 The Author(s).
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: [G. Thomas Caltagirone and Albert Liao reports a relationship with Aptagen that includes: employment.].
Figures


Similar articles
-
Aptamer Sandwich Assay for the Detection of SARS-CoV-2 Spike Protein Antigen.ACS Omega. 2021 Dec 15;6(51):35657-35666. doi: 10.1021/acsomega.1c05521. eCollection 2021 Dec 28. ACS Omega. 2021. PMID: 34957366 Free PMC article.
-
Accessible and Adaptable Multiplexed Real-Time PCR Approaches to Identify SARS-CoV-2 Variants of Concern.Microbiol Spectr. 2022 Oct 26;10(5):e0322222. doi: 10.1128/spectrum.03222-22. Epub 2022 Sep 15. Microbiol Spectr. 2022. PMID: 36106882 Free PMC article.
-
Development of robust, indigenous ELISA for detection of IgG antibodies against CoV-2 N and S proteins: mass screening.Appl Microbiol Biotechnol. 2022 Sep;106(18):6225-6238. doi: 10.1007/s00253-022-12113-8. Epub 2022 Aug 17. Appl Microbiol Biotechnol. 2022. PMID: 35976427 Free PMC article.
-
Aptamers targeting SARS-COV-2: a promising tool to fight against COVID-19.Trends Biotechnol. 2023 Apr;41(4):528-544. doi: 10.1016/j.tibtech.2022.07.012. Epub 2022 Aug 1. Trends Biotechnol. 2023. PMID: 35995601 Free PMC article. Review.
-
Immunologic Testing for SARS-CoV-2 Infection from the Antigen Perspective.J Clin Microbiol. 2021 Apr 20;59(5):e02160-20. doi: 10.1128/JCM.02160-20. Print 2021 Apr 20. J Clin Microbiol. 2021. PMID: 33318065 Free PMC article. Review.
Cited by
-
Optical biosensors for diagnosis of COVID-19: nanomaterial-enabled particle strategies for post pandemic era.Mikrochim Acta. 2024 May 10;191(6):320. doi: 10.1007/s00604-024-06373-6. Mikrochim Acta. 2024. PMID: 38727849 Free PMC article. Review.
-
Development and characterization of high-affinity aptamers for HIV protease detection.Heliyon. 2024 Sep 20;10(22):e38234. doi: 10.1016/j.heliyon.2024.e38234. eCollection 2024 Nov 30. Heliyon. 2024. PMID: 39624307 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
