The Drosophila chemokine-like Orion bridges phosphatidylserine and Draper in phagocytosis of neurons
- PMID: 37276397
- PMCID: PMC10268242
- DOI: 10.1073/pnas.2303392120
The Drosophila chemokine-like Orion bridges phosphatidylserine and Draper in phagocytosis of neurons
Abstract
Phagocytic clearance of degenerating neurons is triggered by "eat-me" signals exposed on the neuronal surface. The conserved neuronal eat-me signal phosphatidylserine (PS) and the engulfment receptor Draper (Drpr) mediate phagocytosis of degenerating neurons in Drosophila. However, how PS is recognized by Drpr-expressing phagocytes in vivo remains poorly understood. Using multiple models of dendrite degeneration, we show that the Drosophila chemokine-like protein Orion can bind to PS and is responsible for detecting PS exposure on neurons; it is supplied cell-non-autonomously to coat PS-exposing dendrites and to mediate interactions between PS and Drpr, thus enabling phagocytosis. As a result, the accumulation of Orion on neurons and on phagocytes produces opposite outcomes by potentiating and suppressing phagocytosis, respectively. Moreover, the Orion dosage is a key determinant of the sensitivity of phagocytes to PS exposed on neurons. Lastly, mutagenesis analyses show that the sequence motifs shared between Orion and human immunomodulatory proteins are important for Orion function. Thus, our results uncover a missing link in PS-mediated phagocytosis in Drosophila and imply conserved mechanisms of phagocytosis of neurons.
Keywords: Orion; da neurons; epidermal cells; phagocytosis; phosphatidylserine.
Conflict of interest statement
The authors declare no competing interest.
Figures
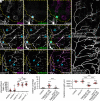

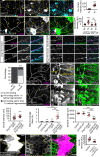




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

