Odd-even disparity in the population of slipped hairpins in RNA repeat sequences with implications for phase separation
- PMID: 37276412
- PMCID: PMC10268303
- DOI: 10.1073/pnas.2301409120
Odd-even disparity in the population of slipped hairpins in RNA repeat sequences with implications for phase separation
Abstract
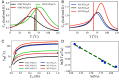
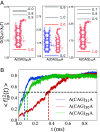
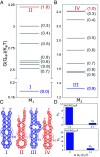
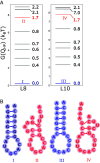
Low-complexity nucleotide repeat sequences, which are implicated in several neurological disorders, undergo liquid-liquid phase separation (LLPS) provided the number of repeat units, n, exceeds a critical value. Here, we establish a link between the folding landscapes of the monomers of trinucleotide repeats and their propensity to self-associate. Simulations using a coarse-grained Self-Organized Polymer (SOP) model for (CAG)n repeats in monovalent salt solutions reproduce experimentally measured melting temperatures, which are available only for small n. By extending the simulations to large n, we show that the free-energy gap, ΔGS, between the ground state (GS) and slipped hairpin (SH) states is a predictor of aggregation propensity. The GS for even n is a perfect hairpin (PH), whereas it is a SH when n is odd. The value of ΔGS (zero for odd n) is larger for even n than for odd n. As a result, the rate of dimer formation is slower in (CAG)30 relative to (CAG)31, thus linking ΔGS to RNA-RNA association. The yield of the dimer decreases dramatically, compared to the wild type, in mutant sequences in which the population of the SH decreases substantially. Association between RNA chains is preceded by a transition to the SH even if the GS is a PH. The finding that the excitation spectrum-which depends on the exact sequence, n, and ionic conditions-is a predictor of self-association should also hold for other RNAs (mRNA for example) that undergo LLPS.
Keywords: RNA–RNA association; excited states; liquid–liquid phase separation; low complexity RNA sequences; self-organized polymer model.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
Salt-Dependent Self-Association of Trinucleotide Repeat RNA Sequences.J Phys Chem Lett. 2024 Apr 11;15(14):3820-3827. doi: 10.1021/acs.jpclett.3c03553. Epub 2024 Apr 1. J Phys Chem Lett. 2024. PMID: 38557079
-
Structure of even/odd trinucleotide repeat sequences modulates persistence of non-B conformations and conversion to duplex.Biochemistry. 2011 May 31;50(21):4441-50. doi: 10.1021/bi200397b. Epub 2011 May 4. Biochemistry. 2011. PMID: 21526744 Free PMC article.
-
Dynamics of strand slippage in DNA hairpins formed by CAG repeats: roles of sequence parity and trinucleotide interrupts.Nucleic Acids Res. 2020 Mar 18;48(5):2232-2245. doi: 10.1093/nar/gkaa036. Nucleic Acids Res. 2020. PMID: 31974547 Free PMC article.
-
Trinucleotide repeats associated with human disease.Nucleic Acids Res. 1997 Jun 15;25(12):2245-54. doi: 10.1093/nar/25.12.2245. Nucleic Acids Res. 1997. PMID: 9171073 Free PMC article. Review.
-
Trinucleotide repeat DNA structures: dynamic mutations from dynamic DNA.Curr Opin Struct Biol. 1998 Jun;8(3):321-30. doi: 10.1016/s0959-440x(98)80065-1. Curr Opin Struct Biol. 1998. PMID: 9666328 Review.
Cited by
-
Stick-slip unfolding favors self-association of expanded HTT mRNA.Nat Commun. 2024 Oct 9;15(1):8738. doi: 10.1038/s41467-024-52764-x. Nat Commun. 2024. PMID: 39384800 Free PMC article.
-
Stick-slip unfolding favors self-association of expanded HTT mRNA.bioRxiv [Preprint]. 2024 Jun 3:2024.05.31.596809. doi: 10.1101/2024.05.31.596809. bioRxiv. 2024. Update in: Nat Commun. 2024 Oct 9;15(1):8738. doi: 10.1038/s41467-024-52764-x. PMID: 38895475 Free PMC article. Updated. Preprint.
-
Sequence complexity and monomer rigidity control the morphologies and aging dynamics of protein aggregates.Proc Natl Acad Sci U S A. 2024 Dec 10;121(50):e2409973121. doi: 10.1073/pnas.2409973121. Epub 2024 Dec 6. Proc Natl Acad Sci U S A. 2024. PMID: 39642206 Free PMC article.
-
Biomolecular condensates control and are defined by RNA-RNA interactions that arise in viral replication.Res Sq [Preprint]. 2025 May 13:rs.3.rs-6378534. doi: 10.21203/rs.3.rs-6378534/v1. Res Sq. 2025. PMID: 40470224 Free PMC article. Preprint.
-
Controlling intermolecular base pairing in Drosophila germ granules by mRNA folding and its implications in fly development.Nat Commun. 2025 Aug 30;16(1):8135. doi: 10.1038/s41467-025-62973-7. Nat Commun. 2025. PMID: 40885704 Free PMC article.
References
-
- Eisenberg H., Felsenfeld G., Studies of temperature-dependent conformation and phase separation of polyriboadenylic acid solution at neutral ph. J. Mol. Biol. 30, 17 (1967). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

