INSurVeyor: improving insertion calling from short read sequencing data
- PMID: 37277343
- PMCID: PMC10241795
- DOI: 10.1038/s41467-023-38870-2
INSurVeyor: improving insertion calling from short read sequencing data
Abstract
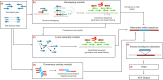
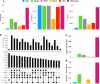
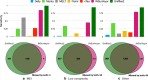
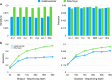
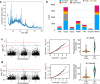
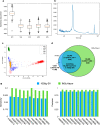
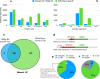
Insertions are one of the major types of structural variations and are defined as the addition of 50 nucleotides or more into a DNA sequence. Several methods exist to detect insertions from next-generation sequencing short read data, but they generally have low sensitivity. Our contribution is two-fold. First, we introduce INSurVeyor, a fast, sensitive and precise method that detects insertions from next-generation sequencing paired-end data. Using publicly available benchmark datasets (both human and non-human), we show that INSurVeyor is not only more sensitive than any individual caller we tested, but also more sensitive than all of them combined. Furthermore, for most types of insertions, INSurVeyor is almost as sensitive as long reads callers. Second, we provide state-of-the-art catalogues of insertions for 1047 Arabidopsis Thaliana genomes from the 1001 Genomes Project and 3202 human genomes from the 1000 Genomes Project, both generated with INSurVeyor. We show that they are more complete and precise than existing resources, and important insertions are missed by existing methods.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Miki Y, et al. Disruption of the apc gene by a retrotransposal insertion of l1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

