Systematically identifying genetic signatures including novel SNP-clusters, nonsense variants, frame-shift INDELs, and long STR expansions that potentially link to unknown phenotypes existing in dog breeds
- PMID: 37277710
- PMCID: PMC10240460
- DOI: 10.1186/s12864-023-09390-6
Systematically identifying genetic signatures including novel SNP-clusters, nonsense variants, frame-shift INDELs, and long STR expansions that potentially link to unknown phenotypes existing in dog breeds
Abstract
Background: In light of previous studies that profiled breed-specific traits or used genome-wide association studies to refine loci associated with characteristic morphological features in dogs, the field has gained tremendous genetic insights for known dog traits observed among breeds. Here we aim to address the question from a reserve perspective: whether there are breed-specific genotypes that may underlie currently unknown phenotypes. This study provides a complete set of breed-specific genetic signatures (BSGS). Several novel BSGS with significant protein-altering effects were highlighted and validated.
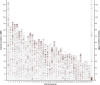
Results: Using the next generation whole-genome sequencing technology coupled with unsupervised machine learning for pattern recognitions, we constructed and analyzed a high-resolution sequence map for 76 breeds of 412 dogs. Genomic structures including novel single nucleotide polymorphisms (SNPs), SNP clusters, insertions, deletions (INDELs) and short tandem repeats (STRs) were uncovered mutually exclusively among breeds. We also partially validated some novel nonsense variants by Sanger sequencing with additional dogs. Four novel nonsense BSGS were found in the Bernese Mountain Dog, Samoyed, Bull Terrier, and Basset Hound, respectively. Four INDELs resulting in either frame-shift or codon disruptions were found in the Norwich Terrier, Airedale Terrier, Chow Chow and Bernese Mountain Dog, respectively. A total of 15 genomic regions containing three types of BSGS (SNP-clusters, INDELs and STRs) were identified in the Akita, Alaskan Malamute, Chow Chow, Field Spaniel, Keeshond, Shetland Sheepdog and Sussex Spaniel, in which Keeshond and Sussex Spaniel each carried one amino-acid changing BSGS in such regions.
Conclusion: Given the strong relationship between human and dog breed-specific traits, this study might be of considerable interest to researchers and all. Novel genetic signatures that can differentiate dog breeds were uncovered. Several functional genetic signatures might indicate potentially breed-specific unknown phenotypic traits or disease predispositions. These results open the door for further investigations. Importantly, the computational tools we developed can be applied to any dog breeds as well as other species. This study will stimulate new thinking, as the results of breed-specific genetic signatures may offer an overarching relevance of the animal models to human health and disease.
Keywords: Dogs as model organism; INDELs; SNP clusters; Short Tandem Repeats; Whole genome sequencing experiments and analyses.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
An autosomal recessive mutation in SCL24A4 causing enamel hypoplasia in Samoyed and its relationship to breed-wide genetic diversity.Canine Genet Epidemiol. 2017 Nov 22;4:11. doi: 10.1186/s40575-017-0049-1. eCollection 2017. Canine Genet Epidemiol. 2017. PMID: 29201383 Free PMC article.
-
Common variants in the CPT1A gene are associated with cataracts in Northern breeds of domestic dog.PLoS One. 2025 Apr 4;20(4):e0320878. doi: 10.1371/journal.pone.0320878. eCollection 2025. PLoS One. 2025. PMID: 40184359 Free PMC article.
-
Prevalence of the breed-related glaucomas in pure-bred dogs in North America.Vet Ophthalmol. 2004 Mar-Apr;7(2):97-111. doi: 10.1111/j.1463-5224.2004.04006.x. Vet Ophthalmol. 2004. PMID: 14982589
-
Forensic DNA phenotyping: Canis familiaris breed classification and skeletal phenotype prediction using functionally significant skeletal SNPs and indels.Anim Genet. 2022 Jun;53(3):247-263. doi: 10.1111/age.13165. Epub 2021 Dec 28. Anim Genet. 2022. PMID: 34963196 Review.
-
Repeat polymorphism analysis for examining genetic variability and the implications for specific traits in dog breeds.Genes Genet Syst. 2013;88(5):273-8. doi: 10.1266/ggs.88.273. Genes Genet Syst. 2013. PMID: 24694390 Review.
References
-
- Marsden CD, Ortega-Del Vecchyo D, O’Brien DP, Taylor JF, Ramirez O, Vilà C, Marques-Bonet T, Schnabel RD, Wayne RK, Lohmueller KE. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc Natl Acad Sci. 2016;113(1):152–157. doi: 10.1073/pnas.1512501113. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources